|
Fibroblast Growth
Factors
&
Their Receptors:
A Kinemage
Collection
by
Larry P. Taylor,
Ph. D. (retired)
Feedback
appreciated; please send comments to:
Email:
lpt
Molecular & Behavioral
Neuroscience Institute
The University of
Michigan
Ann Arbor, MI
This
site is best viewed with
a screen
resolution of 1280 x 1024. |

|
My University Home
Harris
Links Chemistry / Modeling
Links
FGF Site: Downloads
Nomenclature Notes
References FGF
Sequences FGFR
Sequences
Fibroblast Growth
Factors & Their Receptors
Fibroblast Growth Factors (FGF's) represent a homologous family of at least
18 proteins with a beta trefoil (three-fold repeat of a four-stranded sheet assembly without extended alpha helix
strands possessing a pseudo three-fold axis of symmetry) motif associated with cell proliferation and differentiation.
They are a prime component of angiogenesis associated with organogenesis, tumor growth, and wound healing.
Imbalances in FGF levels and/or mutations of the gene encoding for FGF or its
receptors have been associated with numerous diseases, cancers and other
pathological states. Although the FGF ligands share similar sequences and
three-dimensional
motif, there are subtle sequence-dependent backbone
differences than can rationalize
much of the observed FGF to FGF receptor binding behavior. FGF 1 is considered the universal FGF ligand since it binds with high affinity to
all four of the
major human FGF receptors. Each of the FGF ligands is summarized here.
A sub-family of the 23 homologous FGF related proteins, the FHF family (FGF 12
(FHF 1))
of protein ligands was recently removed (lack of functional
similarity) from the FGF family. This change in "status" of the FHF
sub-family of FGF ligands means that the FGF protein family is now composed of at least
18 homologous proteins that bind FGF receptors.
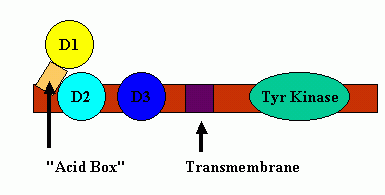
FGF molecules function by interacting with FGF receptors (FGFR's). These receptors typically contain
three Ig-like binding domains (with domains D2 and D3 being most involved in
FGF-FGF receptor binding interactions), a short transmembrane spanning domain and a cytoplasmic component that possesses tyrosine kinase activity.
Between receptor protein Domains D1 and D2 is a short span of acidic residues called the "Acid
Box." A cartoon schematic of FGF Receptor Domains is shown
below:
FGF Receptor Domains.
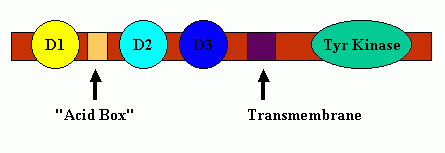
The D1 and the linker region between D1-D2 have much sequence
variability, multiple splice variants, are structurally disordered (even in the presence of bound
FGF ligand), and are considered
not necessary for ligand binding ( FGF binds to FGFR even in the total absence of protein receptor domain
D1). It is
hypothesized that the "Acid Box" can participate in the regulation of FGF binding
to FGFR's in that the multiple acidic sites of the "Acid
Box" can mimic the negative potential
surface of
heparin-like compounds and thus, bind at the heparin-binding
sites located on the surface of the D2 region of the FGF receptor without initiating a biological response (an auto-inhibition mechanism).
A cartoon of this inhibition is shown below:
"Acid Box" Mediated Auto-Inhibition
There are 4 major FGF receptors, but multiple splice variants, especially in
exon III of the receptors (corresponding to FGF receptor protein domain D3) add to the
complexity of the ligand-receptor binding event The third exon contains
three parts: (IIIa, IIIb, and IIIc) and gene splicing events lead to D3 domain transcribed
from the invariant IIIa portion of the gene followed by either b or c.
This splice variation in the ligand binding domain defined by protein receptor
domains D2 and D3 splits the four main receptors into a total of seven key
human FGF receptors: FGFR 1b , FGFR 1c, FGFR 2b , FGFR 2c, FGFR
3b, FGFR 3c, and FGFR 4. The alternative splicing in D3 apparently
is a tissue-specific process with isoform c associated with mesenchymal and
isoform b predominate in epithelial cells.
An essential component of biological activity is the heparin mediated
dimerization of the FGF receptor.
Both FGF
ligands and receptors are characterized by descriptions of their beta sheets. (see nomenclature).
Since chemical structure and physiological function are
intimately related, examination of FGF and FGFR molecular architectures
(especially comparing similar molecules with different physiological responses) may
provide a better understanding of how the FGF ligands interact with their receptors to
provide a biological effect. Although static pictures and FGF
and FGFR Sequence Alignments of a molecular family are helpful,
dynamic images better facilitate delineation of molecular shapes and
ligand-receptor interactions responsible for
biological activity. So, this site uses a protein visualization tool, KiNG , to
facilitate real-time on-line manipulation of kinemage renderings of FGF related
molecules. Kinemages for the molecules discussed and the KiNG
Manual (pdf) and the are also available as separate
downloads for off-line use. These kinemages were prepared from crystal structure coordinates
found in the Brookhaven Data Base (References).
The
real-time visualization using KiNG of the structures on this site requires a
java-enabled (JRE from Java) browser.
Possible
Icons to the left of molecular model image on the download page
| Java Not Activated |
Java Not Activated |
Java Functional |
 |
Blank Area
or message:
Image requires a Java enabled browser
|
 |
| KiNG Inactive |
KiNG Inactive |
KiNG Full Functional |
A single
click on the KiNG logo will launch the appropriate kinemage.
According
to the University of Michigan Web Log:
|
Total
visitors since August, 2005: 78,700 (umich
ceased tracking visits: 8 / 01 / 18)
|
| Site
visitors during the last calendar month: 256 |
| Maximum
Monthly Visitors: 2507 |
Top
Jump To: Intro
Molecules
Discussed
FGF Site: Downloads
Nomenclature Notes
References FGF
Sequences FGFR
Sequences
My University Home
Harris
Links Chemistry / Modeling
Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
The University of Michigan
All Rights Reserved
Supported by the
Pritzker
Neuropsychiatric Disorders Research Consortium, and by NIH
Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1
DA13386 and the Office of Naval Research
(ONR) N00014-02-1-0879 to Huda
Akil & Stanley
J. Watson. at the Molecular
& Behavioral Neuroscience Institute.