| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
|
FGF 10 Bound To The Extracellular Ligand Binding Domain of FGFR 2b by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
FGF 10 Bound To The Extracellular Ligand Binding Domain of FGFR 2b
Most FGF species bind to, and thus activate, multiple FGF receptors. However, the FGF 7 sub-family (FGF 7, 10 and 22) activates only the FGFR 2b splice variant. (FGF receptors 1-3 have an alternative splicing situation in the D3 domain region of the receptor. Here, the D3 domain originates from an invariant exon (a), followed by one of two alternatives (b or c). FGF and FGF 10 both compete for this tissue specific FGFR 2b splice variant. Mouse knock-out studies have established FGF 10 as the primary ligand for the FGF receptor 2b splice variant.
The
unit cell Calpha trace is shown in Kinemage 1. Kinemage
2 adds ribbon rendering, while Kinemage 3 shows the
secondary structure cartoon for the unit cell. The characteristics of the unit
cell for this structure are summarized at pdbsum.
The FGF 10 molecule has three distinguishing characteristics compared to the FGF 1 and FGF 2 template. The most obvious is a N-terminal alpha helix region. This region resides immediately prior to an extended beta sheet 1 (See
Kinemage 4 for comparison of FGF 7 and 10 superimposed on FGF 2) Finally, the FGF 7 sub-family shows an extended turn associated with the low affinity binding site. These features result in a FGF to FGF receptor binding geometry that is different from less selective FGF ligands such as FGF 1 and FGF 2.
The alpha helix N-terminal region has extensive interactions with the D3 region of the FGF 2b splice variant receptor. This is shown in
cartoon format in Kinemage 5 with specific residue interactions highlighted in
Kinemage 6. The ligand-receptor 2b interactions in the D3 region of the FGF receptor are primarily a network of hydrogen bonds. Specifically, Leu-73 (backbone O) to Ser-282 (backbone N), Gln-74 (Oe1) to Lys-279 (side chain N), Gly-75 (backbone N via water) to Val-280 (backbone O), Asp-76 (O d2) to Ala-322 (backbone N), Asp-76 (O d1 and O d2) to Ser-315 (side chain O), Asp-76 (backbone O via water) to Gly-316 (backbone N), Arg-78 (side chain NH2) to Ser-282 (backbone O), Arg-78 (side chain NH2) to Asp-283 (side chain O), Thr-114 (side chain O via water) to Gly-316 (backbone N and O), Glu-154 (side chain O1) to Gln-285 (backbone N) and Glu-154 (side chain O2 via water) to Gln-285 (side chain O1). There is also a hydrophobic contact between ligand residue Phe-146 with receptor amino acid Ile-317.
The ligand to receptor D2-D3 linker region interactions center around the receptor invariant Arg-251 residue. This residue is held in place by a hydrogen bond to receptor residue Asp-283. This sets the Arginine residue in position to hydrogen bond to the backbone carbonyl of
Gly-160 and to form two hydrogen bonds to the carbonyl of Asn-162. In most FGF receptors, the 160 position is
occupied by a relatively bulky hydrogen bonding amino acid (H, N or Y). The Glycine-160 residue found in FGF 10 allows a much closer contact between the FGF 10 ligand and the FGFR 2b receptor linker region. For example, measurement of the Calpha distance between Arg-251 of the receptor and the Calpha of the ligand position 160 is 5.499 angstroms with FGF 10 (bound to FGFR 2b) compared to 6.373 angstroms in FGF 2 (bound to FGFR 2c). This
measurement is shown in Kinemage 7, view 6. Finally, the relative position of the linker region with respect to the receptor D2 and D3 regions is controlled by a Proline residue acting as a hinge. This Pro-253, as in most FGF receptors, is in the trans conformation.
Comparison of FGF 10 bound to FGFR 2c and FGF 2 bound to FGFR 2b shows the positions of FGF and the receptor D3 and D2-D3 linker to be nearly identical. However, the relative position of FGF 10 ligand to D2 is different. This rotation of D2 and the resultant ligand-receptor binding is believed to be ligand induced and the prime component in FGF 7 sub-family preference for the IIIc splice variant of FGFR 2. (This rotation of splice variant c compared to slice variant b can be seen in
Kinemage 5)
The interaction of FGF 7 sub-family ligand with FGF receptors in the D2 region (Kinemage
8 ) has one distinctly critical difference. FGF 1 and 2 have a Tyr residue (position 29 in FGF 1 and position 32 in FGF 3) that is involved in hydrophobic interactions and hydrogen bonding networks with the FGF receptors. In the FGF 7 sub-family, this residue is Phe-83,
which of course, cannot participate in forming a network of hydrogen bonds. Instead, the stabilizing network
of hydrogen bonds is formed with Glu-104 and Asn-151 of the FGF 10 ligand and Leu-166 and Asn-173 of the 2b receptor. There is also an oxy-aromatic interaction between receptor residue Asn-173 and FGF 10 residues Phe-85 and Tyr-161.
Since Asn-173 is conserved only in the FGFR 2 family, the combination of D2 rotation of the FGFR 2 b (relative to FGFR 2 c) and the hydrogen bonding of Asn-173 are considered the major factors in conferring FGF sub-family specificity to the FGFR 2b splice variant.
The Kinemages
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
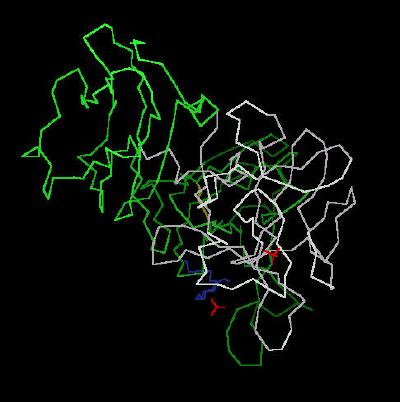
Kinemage 1: The Calpha carbon trace of the ligand receptor complex
View 1 the complex from the crystal
View 2 arbitrary "top view"
View 3 corresponding front view
|
21 K |
 |
| Click on KiNG to see | FGF 10 Bound To FGFR 2b |
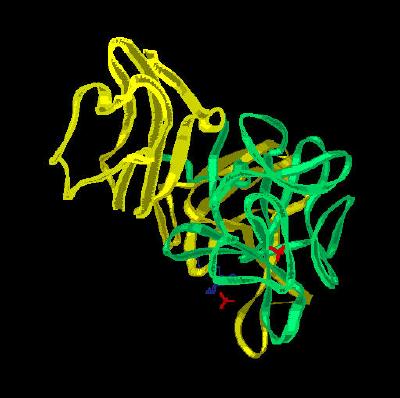
Kinemage 2: The ribbon cartoon rendering of the ligand-receptor complex
View 1 the complex from the crystal
View 2 arbitrary "top view"
View 3 corresponding front view
|
594 K |
 |
| Click on KiNG to see | Cartoon Rendering: FGF 10-FGFR 2b |
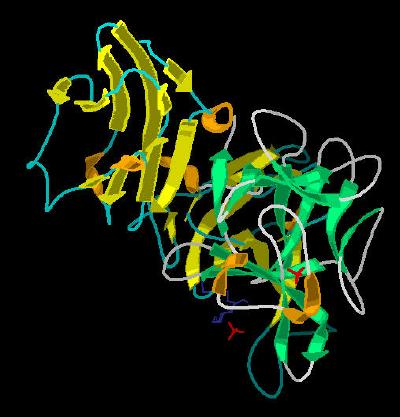
Kinemage 3: The secondary structure cartoon of the ligand receptor complex
View 1 the complex from the crystal
View 2 arbitrary "top view"
View 3 corresponding front view
|
480 K |
 |
| Click on KiNG to see | Secondary Structure Rendering: FGF 10-FGFR 2b |
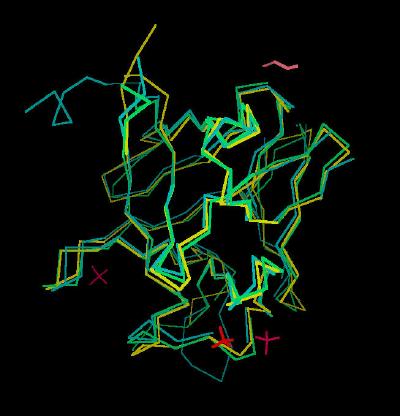
Kinemage 4: A Composite Backbone Overlay of FGF 7 and 10 upon FGF 2
View 1 the overlay
View 2 close up of the N and C terminal regions
View 3 close up of the low affinity loop
|
431 K |
 |
| Click on KiNG to see | FGF 7 & 10 Superimposed Upon FGF 2 |
Kinemage 5: A Composite Backbone Overlay of FGF 10-FGFR 2b upon FGF 2-FGFR 2c
View 1 complex from the crystal
View 2 an arbitrary "top view"
View 3 corresponding front view
View 4 FGF ligand interaction with the D3 region of the FGF receptor
View 5 another view of the FGF ligand interaction with the D3 region of the FGF receptor
|
740 K |
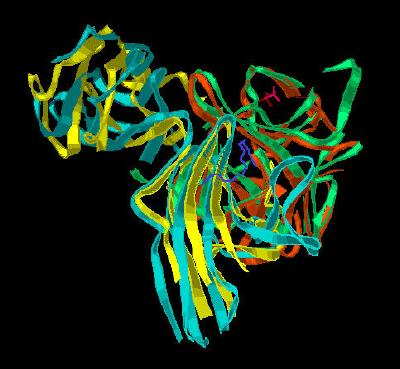 |
| Click on KiNG to see | Overlay of FGF 10-FGFR 2b upon FGF 2-FGFR 2c |
Kinemage 6: The FGF 10 Ligand Interactions with the D3 Domain of FGFR 2b
The ligand-D3 interactions include hydrogen bonds at Leu-73 (backbone O) to Ser-282 (backbone N), Gln-74 (Oe1) to Lys-279 (side chain N), Gly-75 (backbone N via water) to Val-280 (backbone O), Asp-76 (O d2) to Ala-322 (backbone N), Asp-76 (O d1 and O d2) to Ser-315 (side chain O), Asp-76 (backbone O via water) to Gly-316 (backbone N), Arg-78 (side chain NH2) to Ser-282 (backbone O), Arg-78 (side chain NH2) to Asp-283 (side chain O), Thr-114 (side chain O via water) to Gly-316 (backbone N and O), Glu-154 (side chain O1) to Gln-285 (backbone N) and Glu-154 (side chain O2 via water) to Gln-285 (side chain O1).
View 1 the Complex
View 2 arbitrary "top view"
View 3 corresponding front view
View 4 close up of the FGF 10 ligand to FGFR 2b D3 interaction ... arbitrary end view
View 5 close up of the FGF 10 ligand to FGFR 2b D3 interaction ... arbitrary front view
View 6 close up of the FGF 10 ligand to FGFR 2b D3 interaction ... arbitrary opposite end view
View 7 hydrophobic contact site (ligand Phe-146 with receptor Ile-317).
|
514 K |
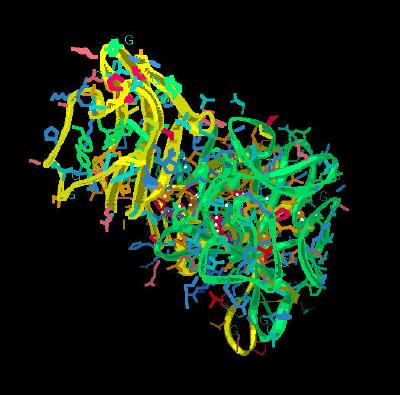 |
| Click on KiNG to see | D3 Domain Interactions for FGF 10-FGFR 2b |
Kinemage 7: The FGF 10 Ligand Interactions with D2-D3 Linker Region of FGFR 3b
The ligand to receptor D2-D3 linker region interactions center around the receptor invariant Arg-251 residue. This residue is held in place by a hydrogen bond to receptor residue Asp-283. This sets the Arginine residue in position to hydrogen bond to the backbone carbonyl of Gly-160 and to form two hydrogen bonds to the carbonyl of Asn-162. In most FGF receptors, the 160 position is occupied by a relatively bulky hydrogen bonding amino acid (H, N or Y). The Glycine-160 residue found in FGF 10 allows a much closer contact between the FGF 10 ligand and the FGFR 2b receptor linker region.
View 1 the complex
View 2 arbitrary "top view"
View 3 corresponding front view
View 4 H bonding of critical linker region residue Arg-251 to the ligand residues Gly-160 (backbone O), 2 hydrogen bonds to the side chain carbonyl of Asn-162 and receptor residue Asp-283.
View 5 Pro-253 in the trans conformation which is the junction between D3 and the linker region
View 6 distance between Calpha's of Arg-251 on the receptor and position 160 of the ligand (toggle measurement on)
|
510 K |
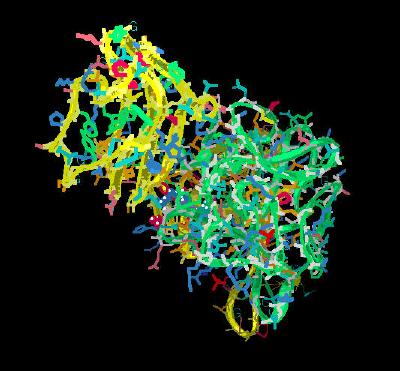 |
| Click on KiNG to see | D2-D3 Linker Interactions for FGF 10-FGFR 2b |
Kinemage 8: The FGF 10 Ligand Interactions with D2 Region of FGFR 3b
The stabilizing network
of hydrogen bonds is formed with Glu-104 and Asn-151 of the FGF 10 ligand and Leu-166 and Asn-173 of the 2b receptor. There is also an oxy-aromatic interaction between receptor residue Asn-173 and FGF 10 residues Phe-85 and Tyr-161.
View 1 the Complex
View 2 arbitrary "top view"
View 3 corresponding front view
View 4 Glu-104 to Leu-166 Hydrogen bond
View 5 Asn-159-Asn-173 Hydrogen Bond
View 6 oxy-Aromatic Interaction
|
511 K |
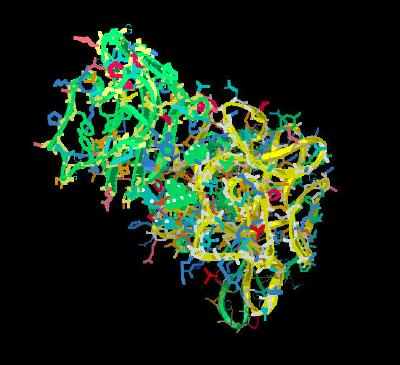 |
| Click on KiNG to see | D2 Domain Interactions for FGF 10-FGFR 2b |
The Sequence
Chain A (FGF 10; residues 69-297)
X-Ray Unresolved N-terminal: GRHVRSYNHLQGDVRWRKLFSFTKYFLKIEKNGKVSGTKKENCPYSILEITSVEIGVVAV
KAIN
X-Ray Resolved: SNYYLAMNKKGKLYGSKEFNNDCKLKERIEENGYNTYASFNWQHNGRQMYVALNGK
GAPRRGQKTRRKNTSAHFLPMVVH
X-Ray Resolved: S
Chain B (FGFR 2 (IIIb) Binding Domain; residues (151-359)
X-Ray Unresolved N-terminal: AEDFVSENSNN
X-Ray Resolved: KRAPYWTNTEKXEKRLHAVPAANTVKFRCPAGGNPXPTXRWLKNGKEFK
QEHRIGGYKVRNQHWSLIXESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQ
AGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINS
SNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPK
X-Ray UnResolved C-terminal: QQAPGREKE
Source:
The sequence of human DNA for residues 64-208 of human FGF 10 was subcloned into a bacterial expression vector pTY-9c. The construct was expressed in Escherichia coli. The sequence of human FGFR 2b residues 140-369 were subcloned in bacterial expression vector pRT-28a. The two proteins were mixed and allowed to crystallize. The structural coordinates are from the Brookhaven Database File1NUN.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.