|
FGF 2-FGFR 1c-Heparin (2:2:2) Complex by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
FGF 2-FGFR 1c-Heparin (2:2:2) Complex
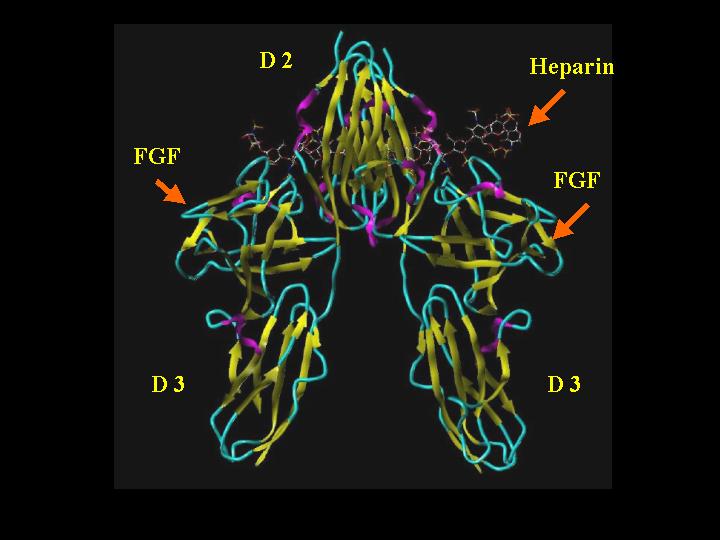
The crystal structure 1FQ9 shows a 2:2:2 arrangement of FGF 2, FGFR 1c, and heparin. This is best considered as 2 separate 1:1 FGF-FGFR complexes that have dimerized via heparin into a complex having a two-fold symmetry. SYBYL cartoon rendering of this complex (Alpha turn = magenta; beta sheet = yellow; coil = cyan; heparin colored by atom type (see table below heparin framework image for standard atom-type coloring) is shown below:
The backbone trace of the unit cell of the 2:2:2 complex is shown in Kinemage
1. Kinemage 2 adds side chains and ribbon rendering, while
Kinemage 3 adds secondary structure cartoon rendering to the unit cell.
The characteristics of the unit cell for this structure are summarized at pdbsum.
Heparin stabilizes of the FGF-FGFR-Heparin complex. In addition to multiple interactions between the FGF ligand and its receptor, heparin also forms hydrogen bonds with another
1:1 FGF-FGFR complex to facilitate dimer formation for an over all FGF-FGFR-Heparin 2:2:2
stoichiometry.
Receptor dimerization is a crucial component of FGF-FGFR interaction and subsequent biological signal activation. The biological process requires heparin sulfate
proteoglycans. Dimerization involves a 1:1 binding of FGF to the D2 and D3 domains of FGF receptors. This 1:1
FGF-FGFR Dimer forms with a cleft or "basic canyon"
(see SYBYL images, below) of positively charged residues in the D2 region of the receptor-ligand interaction volume that would interact with the polyanionic heparin.

FGF 2-FGFR 1c-Heparin (2:2:2) Complex "Front"
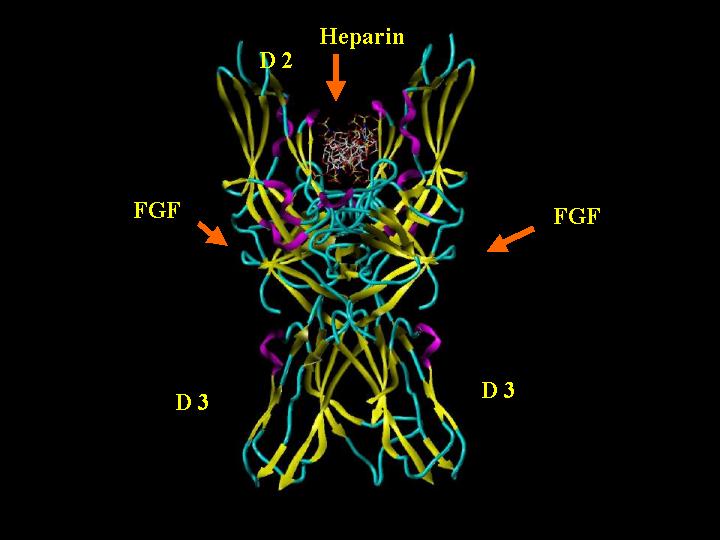
FGF 2-FGFR 1c-Heparin (2:2:2) Complex "Side"
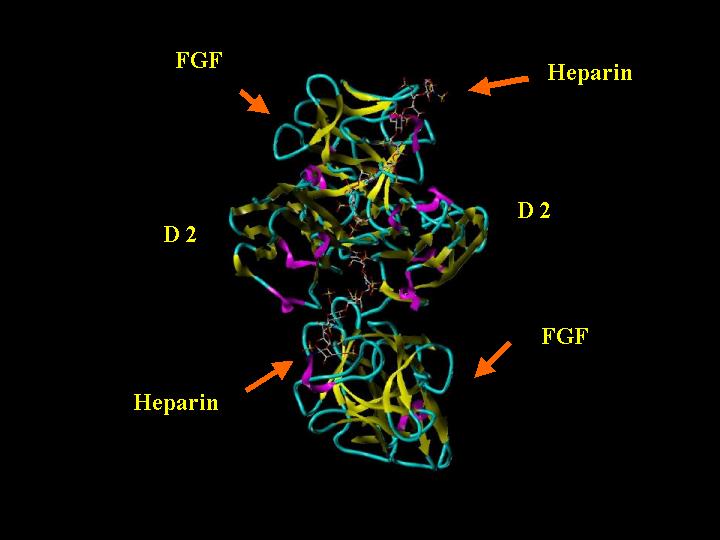
FGF 2-FGFR 1c-Heparin (2:2:2) Complex "Top"
The Heparin decasaccharide in this kinemage is modeled as a helix of repeating disaccharide units of a-D-glucosamine (GlcN) and a-L-iduronic acid (IdoA) joined by alpha-1-4 linkages. Each disaccharide unit has three sulfates: one at the 2-hydroxyl group of IdoA (forming 1,4-dideoxy-O2-sulfo-glucuronic acid ... IDU) and two at the 2-amino and 6-hydroxyl groups of GlcN (forming N,O, 6-disulfo-glucosamine ... SGN). The SYBYL rendering of Chain E from 1FQ9 is shown below
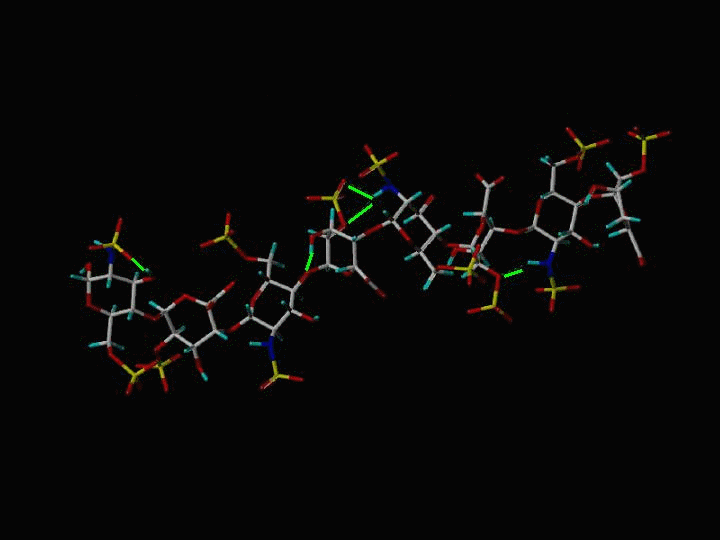
Framework Model of Heparin
|
Atom |
Color |
|
C |
White |
|
H |
Cyan |
|
N |
Blue |
|
O |
Red |
|
S |
Yellow |
|
H-Bonds |
Green Lines |
The sulfate and carboxylate groups form a negative charge potential that lines the ridge of the Heparin helix such that a negative center is found every 17-19 angstroms. The surface potential is shown below. The most negative potential is colored red.
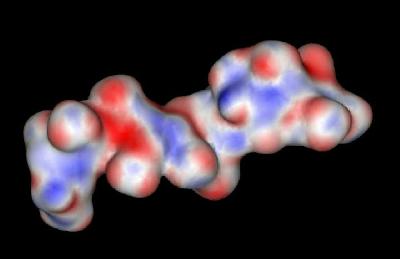
Heparin: Surface Potential
Heparin polysaccharides are quite polar entities with a non-reducing end (O4) and a reducing end (O1). In the crystal structure of the FGF-FGFR-Heparin
(2:2:2) dimer complex, the decasaccharides bind with their non-reducing ends (1,4-dideoxy-5-dehydro-O2-sulfo-glucuronic acid ... UAP) in the center of the FGFR "basic canyon" and continue outward from the interior of the complex onto the high-affinity heparin binding sites of the FGF ligands. This maintains the two-fold symmetry of the dimer complex. (Note that a single, longer
oligomer species of Heparin would not maintain the 2-fold symmetry of the complex).
The glucosamine rings of SGN are all found in a chair conformation. The rings of the other sugars (UAP and IDU) are in either a chair or a skewed boat conformation. This suggests that glucuronic acid ring can adopt multiple orientations depending on the specific FGF or FGFR contact points. This conformational flexibility of the glucuronic acid rings most likely plays a role in specific ligand and/or receptor recognition.
Each carbohydrate ring has both a substructure number assigned by the PDB structural coordinate file and an alphabetic identifier. The correspondence is shown below:
Table 1: PDB Substructure Number vs. Ring Identifier
| Substructure | Ring | Substructure | Ring | |
| 301 | A | 305 | E | |
| 302 | B | 306 | F | |
| 303 | C | 307 | D | |
| 304 | D | 308 | H |
Ring A (Substructure 301) is the non-reducing end of the helix found at the most interior end of
each Heparin chain.
Heparin (designated chains E & F) from the 1FE9 crystal structure is shown in
Kinemage 4.
This kinemage is also available separately (heparin).
There are five intra-molecular hydrogen bonds in Chain E (a 10-mer) and three
intra-molecular hydrogen bonds in chain F (a 6-mer).
The hydrogen bonds between heparin and the D2 Domain of the FGFR 1c are shown in
Kinemage 5. These involve receptor residues Lys-160. Lys-163, Lys-172, Lys-175, and Lys-177. These hydrogen bonds involve all three types (N-sulfate, 2-O-sulfate, and 6-O-sulfate) of heparin sulfate groups
Kinemage 6 shows the interactions between heparin and the FGF Ligand. This involves ligand residues Asn-27, Arg-120, Thr-121, Lys-125, Lys-129, Gln-134, Lys-135, and Ala-136. Except for two hydrogen bonds (one between Lys-135 of FGF 2 and the 6-O-sulfate of ring B and the other between Thr-121 of FGF2 and the 6-O-sulfate group of ring E), the hydrogen bonds between the FGF ligand and heparin involve the heparin N-sulfate and 2-O-sulfate groups. This provides an explanation for FGF2 binding to 6-O-desulfated heparins.
Although most of the heparin interactions are within the FGF-FGFR-heparin monomer, there are some interactions between heparin and the adjacent FGF-FGFR monomer complex to strengthen the stability of the FGF-FGFR-Heparin (2:2:2) dimer.
There are hydrogen bonding interactions involving Lys-207 and Arg-209 and sugar rings
A-D of heparin. In addition, the hydrophobic contacts between Ile-216 and the non reduced ring A of heparin enhance the strength of cross monomer interactions.
The hydrogen bonds between Lys-207 and heparin involve carboxylate, linker, and ring oxygens of heparin.
However, Arg-209 hydrogen bonds with the 2-O-sulfate group of ring C and the 6-O-sulfate group of ring D. This substantiates the dual role of 6-O-sulfate in promoting 1:1 FGF2:FGFR interaction as well as inducing formation of the 2:2 FGF:FGFR dimer. This explains the well-documented inability of 6-O-desulfated heparin oligosaccharides to promote mitogenic activities by failing to induce receptor dimerization. The interactions promoting dimer formation are shown in
Kinemage 7.
Kinemage 8 is a composite of all the heparin interactions with the FGF ligand and its receptor.
Kinemage 9 is an overlap of the FGF-FGFR-heparin dimer (Brookhaven 1FG9) upon the heparin-absent structure (Brookhaven 1CVS). This shows that the addition of heparin does not induce major structural changes in either the FGF ligand or its FGF receptor.
Kinemage 10 highlights the FGF 2 to FGFR 1c Domain 2 interactions. Aside from a hydrogen bond from Tyr-24 of FGF 2 to the receptor backbone at Leu-165, the FGF to receptor interactions that are in the D2 Domain region are primarily hydrophobic. The hydrophobic interactions involve the ligand side chains of Tyr-24 and Met-142 contacting D2 residue Ala-167. Ligand residues Asn-102, Tyr-103, and Leu-140 form a hydrophobic contact with Pro-169 of the D2 Domain of the receptor. Leu-140 also contacts receptor linker region residue Val-248. Since Ala-167, Pro-169, and Val-248 of D2 are well conserved among the four mammalian FGF receptors, the hydrophobic surface of this volume of the receptor may represent a conserved interaction site.
The Ala-167 of FGFR 1c, which interacts with FGF 2 hydrophobic residues, is part of the HAV triad that is the hallmark of cell adhesion molecules.
While the Linker region between D2 and D3 is short, it is well conserved and thus, highly significant in ligand
to receptor binding. Arg-250 is invariant across the four FGF receptors. This Arg residue has its position maintained for ligand interaction by a network of hydrogen bonds that involves both D2 and D3 regions of the receptor. This network provides a fairly rigid and conserved ligand-receptor binding geometry. The guanidinium group of Arg-250 makes two hydrogen bonds with FGF2: one with the backbone carbonyl oxygen of Asn-102 and another with the side chain carbonyl oxygen of Asn-104. An Asn-104 side chain amide in FGF 2 is also engaged in an intra-molecular hydrogen bond with Tyr-106, which in turn is hydrogen bonded to Glu-96, another invariant residue in the FGF ligand family. The side chain of Glu-96 is also hydrogen bonded to the backbone amide nitrogen of Gln-284 in the D3 domain of the receptor. The FGF 2 to
FGFR 1c linker binding interactions are shown in
Kinemage 11.
In addition, the side chains of Val-248 in the D2-D3 linker and the Leu-98 of FGF 2 form a hydrophobic shelf for the aliphatic portion of the Arg-250 side chain. This, plus, the two intra-molecular hydrogen bonds between the Arg-250 guanidinium group and the side chain carboxylate of Asp-282 in D3
(Kinemage 11) may provide a stabilized D2-Linker-D3 region to facilitate ligand binding.
The interactions between D3 and FGF 2 ( Kinemage 12 ) occur near the linker region of the receptor. The side chain of Phe-17 in FGF 2 inserts into a shallow hydrophobic pocket in the
FGFR 1c D3 Domain formed by Pro-285, Ile-287, and the aliphatic portions of the Glu-324 and Asp-320. Also, the back-bone of Phe-17 hydrogen bonds to the Gln-284. The side chain of Lys-21 hydrogen bonds with the side chain of Gln-284 and the backbone of Asp-282.
Residues 56-60 of FGF 2 are in the vicinity of the FGFR 1c D3 Domain and also contribute to ligand binding. The side chain of Gln-56 makes two hydrogen bonds with Asp-320 of D3; one with its backbone amide nitrogen and another with the side chain carboxylate group. In addition, the aliphatic portion of the Gln-56 side chain is in van der Waals contact with Gly-315. Ala-57 hydrophobically interacts with Pro-285 and Gly-315. Glu-58 hydrogen bonds to the carboxylate of the backbone amide of Val-316 and forms hydrophobic contacts with His-286 and Gly-315. The backbone of Glu-59 is in van der Waals contact with the side chain of His-286. The guanidinium group of Arg-60 hydrogen bonds to the backbone carbonyl oxygen of Asn-345. This region of FGF 2 (residues 56-60) is divergent among the 23 members of the FGF family members, suggesting that these residues may confer a significant component of specificity to FGF binding to its receptors.
Without the ligand, Val-316 of the FGFR 1c D3 Domain would be exposed to solvent. However, when ligand-bound, Val-316 hydrophobically interacts with Val-88 of FGF 2. Mutagenesis studies suggest this is an important contact point.
Kinemage 13 highlights the receptor to receptor residues that are important in ligand-receptor complex dimerization. The primary region of receptor to receptor contact is the D2 Domain. Across a 2-fold axis of the dimer, the Ala-171 side chain forms a hydrophobic contact with the corresponding Ala-171 side chain from the other receptor. Ala-171 and Lys-172 atoms are in van der Waals contact with the corresponding atoms of the adjacent receptor. Hydrogen bonds between the side chain of Thr-173 and the backbone nitrogen of Thr-173 of the other receptor and salt bridges between the side chains of Lys-172 and Asp-218 stabilize the interface.
Kinemage 14 highlights the ligand-receptor interactions important for dimerization. In general, these interactions are neither specific nor conserved. The one interaction worth noting is a backbone-to-backbone hydrogen bond between ligand Pro-132 and Gly-204 of the receptor. There are also hydrophobic contact points at the beta
8-9 loop residues Glu-99, Ser-100, Asn-101 and the beta 10-11 loop residues Pro-132, Gly-133, Leu-138 with receptor D3
Domain residues Pro-199, Asp-200, Ile-203, Gly-204, Gly-205, Ser-219, and Val-221.
The side chain of Lys-26 in FGF 2 forms a salt bridge with the side chain of Asp-218 of the D2 Domain of the receptor.
Kinemage 15 shows the backbone superimposition of FGF 2 bound to
FGFR 1c in 1FQ9 and unbound (from Brookhaven 1BFF). The primary difference appears to be in the secondary binding site around the Ser-Asn-Asn sequence. The
small differences in conformation are best seen by toggling on the side chains.
The Kinemages
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1: The Unit Cell for the FGF 2-FGFR 1c-Heparin (2:2:2) Complex
|
65 K |
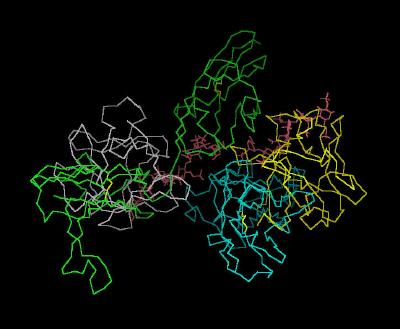 |
| Click on KiNG to see | FGF 2-FGFR 1c-Heparin (2:2:2) Complex |
Kinemage 2: Ribbon Rendering for the FGF 2-FGFR 1c-Heparin (2:2:2) Complex
|
1,134 K |
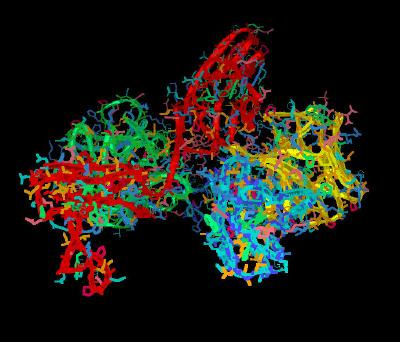 |
| Click on KiNG to see | FGF 2-FGFR 1c-Heparin (2:2:2) Ribbons |
Kinemage 3: Cartoon Rendering for the FGF 2-FGFR 1c-Heparin (2:2:2) Complex
|
911 K |
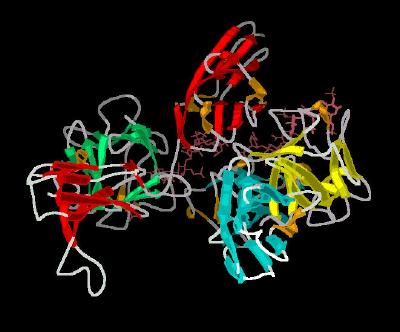 |
| Click on KiNG to see | FGF 2-FGFR 1c-Heparin (2:2:2) Cartoon |
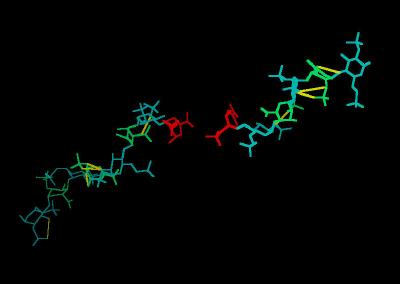
Kinemage 4: Heparin From 1FQ9 Crystal
In the crystal structure of the FGF-FGFR-Heparin (2:2:2) dimer complex, the decasaccharides bind with their non-reducing ends (1,4-dideoxy-5-dehydro-O2-sulfo-glucuronic acid ... UAP) in the center of the FGFR "basic canyon" and continue outward from the interior of the complex onto the high-affinity heparin binding sites of the FGF ligands
The Heparin decasaccharide shown below is modeled as a helix of repeating disaccharide units of a-D-glucosamine (GlcN) and a-L-iduronic acid (IdoA) joined by a-1-4 linkages. Each disaccharide unit has three sulfates: one at the 2-hydroxyl group of IdoA (forming 1,4-dideoxy-O2-sulfo-glucuronic acid ... IDU) and two at the 2-amino and 6-hydroxyl groups of GlcN (forming N,O, 6-disulfo-glucosamine ... SGN).
View 1 Heparin
View 2 intramolecular H-bond Chain E 302-303
View 3 intramolecular H-bond Chain E 304-305 (to both sugar O & sulfate O)
View 4 intramolecular H-bond Chain E 305-306
View 5 intramolecular H-bond Chain E 308-308
View 6 intramolecular H-bond Chain F 302-303
View 7 intramolecular H-bond Chain F 304-305
|
26 K |
 |
| Click on KiNG to see | Heparin from 1FQ9 |
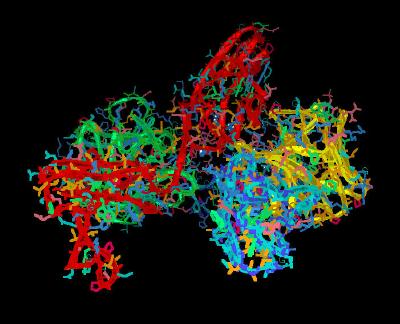
Kinemage 5: Heparin H-bonding to the D2 Domain of the FGF receptor
Heparin interacts with receptor residues Lys-160. Lys-163, Lys-172, Lys-175, and Lys-177. These hydrogen bonds involve all three types (N-sulfate, 2-O-sulfate, and 6-O-sulfate) of heparin sulfate groups.
View 1 the FGF-FGFR-Heparin Complex
View 2 heparin interaction to Lys-160, Chain C
View 3 heparin interaction to Lys-163, Chain C
View 4 heparin interaction to Lys-172, Chain C
View 5 heparin interaction to Lys-175, Chain C
View 6 heparin interaction to Lys-177, Chain C
View 7 "overhead" view of the heparin-receptor interactions at Chain C
View 8 heparin interaction to Lys-160, Chain D
View 9 heparin interaction to Lys-163, Chain D
View 10 heparin interaction to Lys-172, Chain D
View 11 heparin interaction to Lys-175, Chain D
View 12 heparin interaction to Lys-177, Chain D
View 13 "overhead" view of the heparin-receptor interactions at Chain D
|
977 K |
 |
| Click on KiNG to see | Heparin Binding to Receptor D2 Domain |
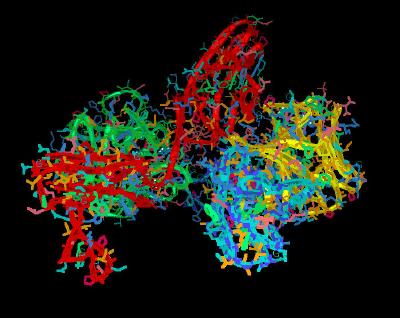
Kinemage 6: Heparin H-bonding to the FGF Ligand
Heparin H-bonding to the FGF Ligand involves ligand residues Asn-27, Arg-120, Thr-121, Lys-125, Lys-129, Gln-134, Lys-135, and Ala-136. Except for two hydrogen bonds (one between Lys-135 of FGF 2 and the 6-O-sulfate of ring B and the other between Thr-121 of FGF2 and the 6-O-sulfate group of ring E), the hydrogen bonds between the FGF ligand and heparin involve the heparin N-sulfate and 2-O-sulfate groups. This provides an explanation for FGF2 binding to 6-O-desulfated heparins.
View 1 the FGF-FGFR-Heparin Complex
View 2 heparin interaction to Asn-27, Chain A
View 3 heparin interaction to Lys-119, Chain A
View 4 heparin interaction to Arg-120, Chain A
View 5 heparin interaction to Lys-125, Chain A
View 6 heparin interaction to Lys-129, Chain A
View 7 heparin interaction to Gln-134, Chain A
View 8 heparin interaction to Asn-27, Chain B
View 9 heparin interaction to Arg-120, Chain B
View 10 heparin interaction to Asn-121, Chain B
View 11 heparin interaction to Lys-125, Chain B
View 12 heparin interaction to Lys-129, Chain B
View 13 heparin interaction to Gln-134, Chain B
|
976 K |
 |
| Click on KiNG to see | Heparin H-bonding to the FGF Ligand |
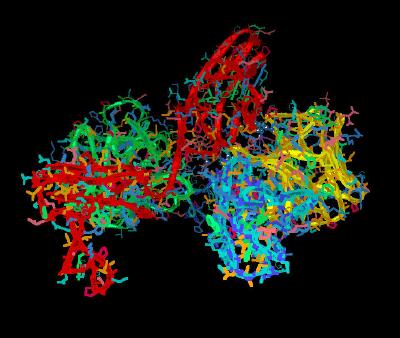
Kinemage 7: Heparin FGF-FGFR Dimer Stabilization
There are hydrogen bonding interactions involving Lys-207 and Arg-209 and sugar rings A-D of heparin. In addition, the hydrophobic contacts between Ile-216 and the non reduced ring A of heparin enhance the strength of cross monomer interactions.
The hydrogen bonds between Lys-207 and heparin involve carboxylate, linker, and ring oxygens of heparin. However, Arg-209 hydrogen bonds with the 2-O-sulfate group of ring C and the 6-O-sulfate group of ring D. This substantiates the dual role of 6-O-sulfate in promoting 1:1 FGF2:FGFR interaction as well as inducing formation of the 2:2 FGF:FGFR dimer. This explains the well-documented inability of 6-O-desulfated heparin oligosaccharides to promote mitogenic activities by failing to induce receptor
dimerization.
View 1 the FGF-FGFR-Heparin Complex
View 2 heparin interaction to Lys-207, Chain D
View 3 heparin interaction to Arg-209, Chain D
View 4 heparin interaction to Lys-207, Chain C
View 5 heparin interaction to Arg-209, Chain C
View 6 heparin interaction to Ile-216, Chain C
View 7 heparin interaction to Ile-216, Chain D
|
973 K |
 |
| Click on KiNG to see | Acidic FGF Receptor Binding Sites |
Kinemage 8: Composite of All the Heparin Interactions
View 1 the FGF-FGFR-Heparin Complex
View 2 heparin interaction to Lys-160, Chain C
View 3 heparin interaction to Lys-163, Chain C
View 4 heparin interaction to Lys-172, Chain C
View 5 heparin interaction to Lys-175, Chain C
View 6 heparin interaction to Lys-177, Chain C
View 7 "overhead" view of the heparin-receptor interactions at Chain C
View 8 heparin interaction to Lys-160, Chain D
View 9 heparin interaction to Lys-163, Chain D
View 10 heparin interaction to Lys-172, Chain D
View 11 heparin interaction to Lys-175, Chain D
View 12 heparin interaction to Lys-177, Chain D
View 13 "overhead" view of the heparin-receptor interactions at Chain D
View 14 heparin interaction to Asn-27, Chain A
View 15 heparin interaction to Lys-119, Chain A
View 16 heparin interaction to Arg-120, Chain A
View 17 heparin interaction to Lys-125, Chain A
View 18 heparin interaction to Lys-129, Chain A
View 19 heparin interaction to Gln-134, Chain A
View 20 heparin interaction to Asn-27, Chain B
View 21 heparin interaction to Arg-120, Chain B
View 22 heparin interaction to Asn-121, Chain B
View 23 heparin interaction to Lys-125, Chain B
View 24 heparin interaction to Lys 129, Chain B
View 25 heparin interaction to Gln-134, Chain B
View 26 heparin interaction to Lys-207, Chain D
View 27 heparin interaction to Arg-209, Chain D
View 28 heparin interaction to Lys-207, Chain C
View 29 heparin interaction to Arg-209, Chain C
View 30 heparin interaction to Ile-216, Chain C
View 31 heparin interaction to Ile-216, Chain D
These views show the canyon cleft created by FGF and FGR that is occupied with Heparin
View 32 looking over Chain B into the cleft
View 33 looking "overhead" at the junction between Heparin Chains E & F
View 34 looking over Chain A into the cleft
View 35 another "overhead" look at the junction between Heparin Chains E & F
|
1,151 K |
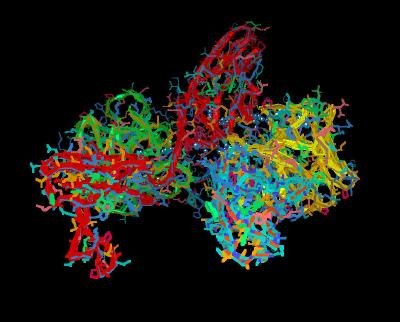 |
| Click on KiNG to see | Composite of all Heparin Binding |
Kinemage 9: Overlap of the FGF-FGFR-Heparin Dimer (Brookhaven 1FG9) upon the Heparin-Absent Structure (Brookhaven 1CVS)
|
1,741 K |
|
| Click on KiNG to see | Overlap of 1FQ9 upon 1CVS |
Kinemage 10: Ligand-D2 Receptor Interactions
A hydrogen bond from Tyr-24 of FGF 2 to the receptor backbone at Leu-165, hydrophobic interactions Tyr-24 and Met-142 contacting D residue Ala-167. Ligand residues Asn-102, Tyr-103, and Leu-140 form a hydrophobic contact with Pro-169 of the D2 Domain of the receptor. Leu-140 also contacts receptor linker region residue Val-248.
View 1 the Complex
View 2 Tyr-24 H bond A
View 3 hydrophobic 1 A
View 4 hydrophobic 2 A
View 5 hydrophobic 3 A
View 6 Tyr-24 H bond B
View 7 hydrophobic 1 B
View 8 hydrophobic 2 B
View 9 hydrophobic 3 B
|
975 K |
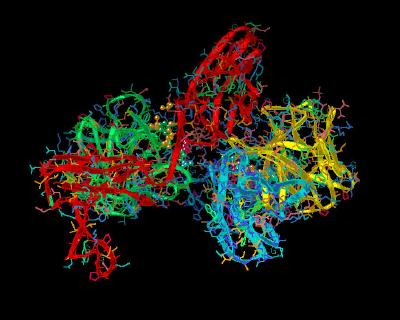 |
| Click on KiNG to see | Ligand-D2 Interactions |
Kinemage 11: Ligand-Linker Receptor Interactions
Arg-250 hydrogen bonds with FGF2: one with the backbone carbonyl oxygen of Asn-102 and another with the side chain carbonyl oxygen of Asn-104. An Asn-104 side chain amide in FGF 2 is also engaged in an intra-molecular hydrogen bond with Tyr-106, which in turn is hydrogen bonded to Glu-96, another invariant residue in the FGF ligand family. The side chain of Glu-96 is also hydrogen bonded to the backbone amide nitrogen of Gln-284 in the D3 domain of the receptor.
View 1 the Complex
View 2 hydrophobic A
View 3 H Bonding 1 A
View 4 H Bonding 2 A
View 5 hydrophobic B
View 6 H Bonding 1 B
View 7 H Bonding 2 B
|
975 K |
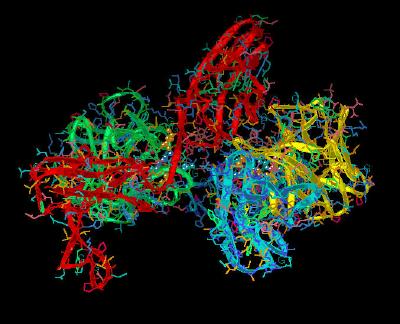 |
| Click on KiNG to see | Ligand-Linker Receptor Interactions |
Kinemage 12: Ligand-D3 Receptor Interactions
Phe-17 in FGF 2 inserts into a shallow hydrophobic pocket in the FGFR 1c D3 Domain formed by Pro-285, Ile-287, and the aliphatic portions of the Glu-324 and Asp-320; back-bone of Phe-17 hydrogen bonds to the Gln-284. The side chain of Lys-21 hydrogen bonds with the side chain of Gln-284 and the backbone of Asp-282. Gln-56 makes two hydrogen bonds with Asp-320 of D3; one with its backbone amide nitrogen and another with the side chain carboxylate group. In addition, the aliphatic portion of the Gln-56 side chain is in van der Waals contact with Gly-315. Ala-57 hydrophobically interacts with Pro-285 and Gly-315. Glu-58 hydrogen bonds to the carboxylate of the backbone amide of Val-316 and forms hydrophobic contacts with His-286 and Gly-315. The backbone of Glu-59 is in van der Waals contact with the side chain of His-286. The guanidinium group of Arg-60 hydrogen bonds to the backbone carbonyl oxygen of Asn-345. Without the ligand, Val-316 of the FGFR 1c D3 Domain would be exposed to solvent. However, when ligand-bound, Val-316 hydrophobically interacts with Val-88 of FGF 2. Mutagenesis studies suggest this is an important contact point.
View 1 the Complex
View 2 Phe-17 Insert A
View 3 H Bonding 1 A
View 4 H Bonding 2 A
View 5 H Bonding 3 A
View 6 V316-V88 (CA)
View 7 Phe-17 Insert B
View 8 H Bonding 1 B
View 9 H Bonding 2 B
View 10 H Bonding 3 B
View 11 V316-V88 (DB)
|
983 K |
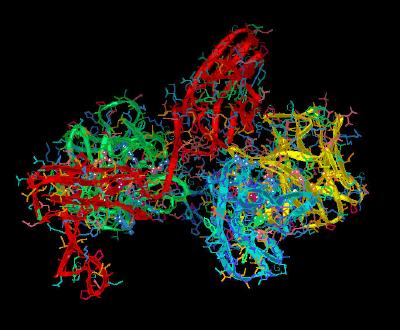 |
| Click on KiNG to see |
Ligand-D3 Interactions |
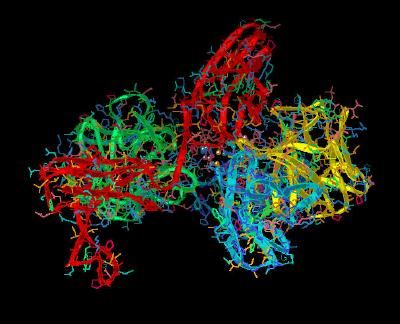
Kinemage 13: Receptor-Receptor Dimerization Sites
Ala-171 and Lys-172 atoms are in van der Waals contact with the corresponding atoms of the adjacent receptor. Hydrogen bonds between the side chain of Thr-173 and the backbone nitrogen of Thr-173 of the other receptor and between the side chains of Lys-172 and Asp-218 stabilize the interface.
View 1 the Complex
View 2 the Dimerization Interface
View 3 H Bonds
|
972 K |
 |
| Click on KiNG to see |
Receptor-Receptor Dimerization Sites: |
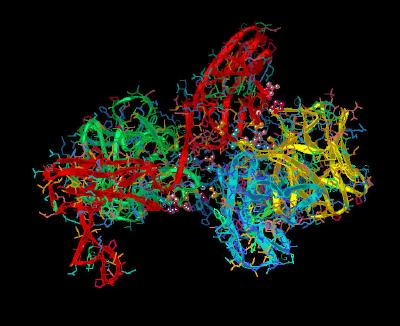
Kinemage 14: Ligand-Receptor Dimerization Sites
There is a hydrogen bond between ligand Pro-132 and Gly-204 of the receptor. There are also hydrophobic contact points at the beta 8-9 loop residues Glu-99, Ser-100, Asn-101 and the beta 10-11 loop residues Pro-132, Gly-133, Leu-138 with receptor D2 Domain residues Pro-199, Asp-200, Ile-203, Gly-204, Gly-205, Ser-219, and Val-221.The side chain of Lys-26 in FGF 2 forms a hydrogen bond with the side chain of Asp-218 of the D2 Domain of the receptor. Since Lys-26 may be a heparin-binding residue, the here-observed interaction with Asp-218 may or may not occur in the presence of heparin.
View 1 the Complex
View 2 Pro-Gly Interaction Chains A & D
View 3 hydrophobic Interactions Chains A & D
View 4 Lys-Asp Salt Bridge Chains A & D
View 5 Pro-Gly Interaction Chains B & C
View 6 hydrophobic Interactions Chains B & C
View 7 Lys-Asp Salt Bridge Chains B & C
|
976 K |
 |
| Click on KiNG to see |
Ligand-Receptor Dimerization Sites: |
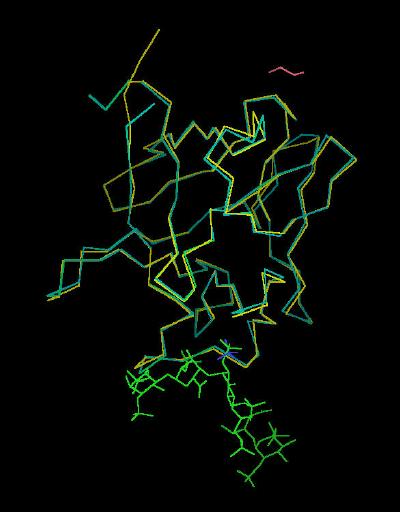
Kinemage 15: Bound vs. Unbound FGF 2 ligand
View 1 FGF 2 from 1FQ9 Chains A & B superimposed on the backbone of FGF 2
View 2 low affinity (discrimination) loop.
View 3 looking directly into the heparin binding region (the "basic Canyon")
View 4 N & C terminal junction
View 5 structural changes upon binding (Ser-100, Asn-101, and Asn-102.)
|
430 K |
 |
| Click on KiNG to see |
FGF 2: Bound vs. Unbound |
Sequences:
Chains A & B: Residues 16-144 of Human FGF 2
Chains C & D: Residues 149-359 of human FGFR 1c
Chain A:
GHFKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYLAMKEDGRLLASKSVTDECFF
FERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQKAILFLPMSAKS
Chain B
GHFKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYLAMKEDGRLLASKSVTDECFF
FERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQKAILFLPMSAKS
Chain C
TDNTKPNRMPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPQPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSV
VPSDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKI
GPDNLPYVQILKTAGVNTTDKEMEVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEER
Chain D
TDNTKPNRMPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPQPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSV
VPSDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKI
GPDNLPYVQILKTAGVNTTDKEMEVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEER
Source:
Existing crystals containing a FGF 2-FGFR 1c dimer (corresponding to Brookhaven Structure 1CVS) were incubated with the heparin decasaccharide. Structural coordinates of the 2:2:2 complex were taken from Brookhaven Database File1FQ9.
Top
FGF Site: FGF Intro
Nomenclature Notes
References FGF
Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
The University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.