
|
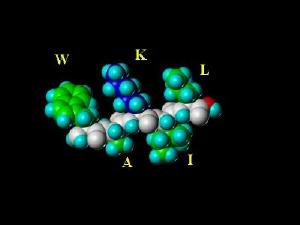
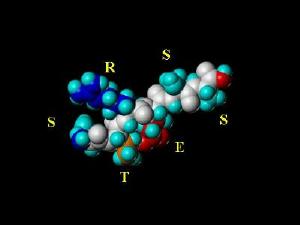
MHRI (in the Amino Acid Font) |
The University Of Michigan Home Page for Larry P. Taylor, Ph. D. (retired) aka
"Harris"
Email:
U of Michigan: lpt Diving: divegeek
This site is best viewed with a display of 1280 x 1024 |
Michigan (in the Amino Acid Font) |
My Michigan Personal Pages : Brain Awareness FGF Kinemages MR Receptor Divegeek Harris Links
My Intro to chemistry web site: funchemistry
Jump On This Page To: Molecular Modeling Fun Chemistry Scuba Diving Web Links Distributed Computing
My background is primarily in organic chemistry. After obtaining my PH.D. in biochemistry while working full-time running a peptide synthesis laboratory, I came to the University of Michigan in 1986 for post-doctoral study in the laboratory of Huda Akil and Stan Watson. I have been with them at MHRI (now the Molecular & Behavioral Neuroscience Institute; MBNI) ever since. Their laboratory is involved in the anatomical and biochemical mapping of the brain circuitry involved in emotional responses, especially stress-related matters. My primary role is to examine molecules involved in these processes. I call it "molecular psychology" (If you were a ligand, where would you be most comfortable?). Since the labs of Huda Akil and Stan Watson are intimately related, those working in their lab have adapted a common name; we refer to the lab as the WAKIL group.
 |
 |
|
The Peptide Images On this Page are chemically correct representations of the amino acid sequences listed in the image (using the standard single letter amino acid code). The peptides were generated with SYBYL molecular modeling software and energy minimized using the Kollman All-Atom Force Field (with consideration of Kollman charges). The coloring scheme (except for school colors used in the MICHIGAN peptide) are based on physical properties of the individual amino acids. I have left the MHRI peptide here since there can be no named peptide for MBNI as there is no "B" in the amino acid code.
Brain Awareness Week Exhibit: Molecular Shape Determines Biological Activity (40 images, ~3.4 meg)
Fibroblast Growth Factors & Their Receptors: A Kinemage Collection
Modeling The MR Receptor (Birkbeck Protein Structure Course Project)
Simple Molecular Modeling (81 images, ~ 4.4 meg)
My Introduction to Chemistry Course Web Site

Click on Start Here to enter the site
Scuba Diving (Diving Myths & Realities)
 |
Has been on-line since 2001
|
 |
Harris Links is my entire collection of bookmarks ( Last Addition: 12 / 11 / 2024).
This compilation is typically downloaded more than 1000 times / month (maximum monthly download: 15,390).
Harris
Links
(
~ 9100 links; ~ 1000
k)
Subsets: Chemistry / Molecular Modeling (~ 800 links; 150 k) Scuba Diving (~1200 links; 145 k)
Classic SETI at Home

Classic SETI (Search for Extraterrestrial Intelligence) @ home was a global collection of people and institutions who donated time on their computers to assist in analyzing radio astronomy data with the hopes that embedded in stellar radio frequency radiation are signals that originate from sentient beings. This involves mathematical signal analysis of enormous proportions. SETI at Home pioneered the process (Distributed Computing) of splitting large computational workloads into small chunks for processing by home computers; managing downloads of data and uploads of finished computations, and finally reassembling and validating results from millions of individual computers to yield a finished determination. This made the SETI at Home project the equivalent of a massive (more than 5.4 million contributors) parallel processing super computer. It is perhaps the best-ever example of true global cooperation. The project terminated in December of 2005. At its termination, my contribution (from May, 1999 to December, 2005) was 17,171 work units (The equivalent of 14.13 years of cpu time). This was more than 99.72 % of all participants.
BOINC
Classic SETI at home evolved into BOINC (Berkeley Open Infrastructure for Network Computing) that uses a common client to manage multiple distributed computing projects. Users can contribute computer processing time to a variety of projects including molecular protein folding, climate analysis, stellar gravitation and several projects directly related to medical research. Researchers can now consider computationally intense problems far beyond the cost and computing power of any single laboratory. Historically, SETI@Home (no new work units as of 3/31/2020 ... officially the project is not abandoned, but in hibernation) was my primary project with Einstein@Home (Gravitational wave detection) as my secondary project. I also contributed to the now retired Predictor at home (Predicting protein 3D structure from an amino acid sequence). In order to keep my idle machines busy when my primary projects have no work available, I also contribute to MilkyWay@Home (Building 3-D map of the Milky Way Galaxy), the now inactive Docking@Home (Ligand Interactions with Proteins) , Rosetta@home (examination of 3-D shape of proteins), and CAS at Home (Predicting Protein Structure).
I was the SETI at Home "User of the day" for December 12, 2008.
Currently Top U of Michigan Contributor
My Detailed BOINC Stats: SETI Einstein MilkyWay Rosetta BOINC Totals
U of MI Team Stats: SETI Einstein MilkyWay Rosetta Total Member Contributions Team Projects
Jump On This Page To: Molecular Modeling Fun Chemistry Scuba Diving Web Links Distributed Computing
My Michigan Personal Pages : Brain Awareness FGF Kinemages MR Receptor Divegeek Harris Links
My Intro to chemistry web site: funchemistry
Contents of this Web Site Copyright 2001- 2025 by Larry "Harris" Taylor, Ph. D.
All
rights reserved.