| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
|
FGF 2 Bound To The Extracellular Ligand Binding Domain of FGFR 2c by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
FGF 2 Bound To The Extracellular Ligand Binding Domain of FGFR 2c
The unit cell of the ligand (FGF 2)-receptor (FGFR 2c) dimer complex is shown in Kinemage 1 as a dimer of dimers. A single dimer component of this complex is visualized in Kinemage 2 (Calphas) , Kinemage 3 (Calphas) , & Kinemage 4 (ribbons). The characteristics of the unit cell for this structure are summarized at pdbsum.
The ligand binding domain of the FGF Receptors is found in the highly conserved immunoglobulin-like regions of the second (D2) and third (D3) domains of the receptor sequence. The primary binding site of the receptor (with the ligand high affinity region) is the highly conserved portion of the D2 domain
(Kinemage 5 ) and the linker between the D2 and D3 FGF receptor
domains (Kinemage 6 ). Ligand specificity is acquired via sequence variability and alternative splicing of the receptor that leads to a highly variable realm within 2
of the loops within the D3 region of the FGF receptor that interacts with the N-terminal and low-affinity loop portions of the FGF ligand. (The D3 Domain is assembled from three exons termed a, b and c. Typically, the
FGFR 2c receptors are composed from invariant exon a with either the b or c exon for the later loops of the D3 Domain. The
D3 region of this receptor is assembled from the exons a and c.)
The high affinity binding region is primarily a site dominated by hydrophobic interactions as the solvent exposed hydrophobic surfaces of both ligand and receptor interact to exclude polar solvent. This is highlighted by FGF residues Tyr-24, Leu-140 and Met-142 packing around Ala-168 of the D2 domain of
FGFR 2c (Kinemage 5). Additionally, Leu-140, Tyr-103 and the aliphatic portion of Asn-102 define a hydrophobic contact volume with Pro-170 of
FGFR 2c. Further interactions suggested by the crystal structure include H bonding between the Tyr-24 hydroxyl of FGF 2 and the backbone of Leu-166 and Ala-168 of
FGFR 2c. There is also a water mediated hydrogen bond between the hydroxyl of Tyr-103 of FGF 2 and the backbone of Ala-168 of
FGFR 2c.
The crystal structure inferences are substantiated with Ala replacement mutations. Separately replacing Tyr-24, Tyr-103, Leu-140, or Met-142 results in substantial decreases in binding affinity between FGF2 and
FGFR 2c.
Another hydrophobic center of interaction occurs between the ligand FGF 2 and the linker region (between D2 and D3) of
FGFR 2c ( Kinemage 6 ) . Hydrophobic interactions provide a distinctly non-polar local environment which will increase any possible electronic interactions. So, the hydrogen bonds in this region between the invariant Arg-253 of the
FGFR 2c receptor with the backbone of Asn-102 and the side chain of Asn-104 of the FGF 2 ligand
are particularly important. Indeed, Ala replacement of FGF 2 Asn-104 yields a 400-fold reduction in binding affinity between FGF 2 and
FGFR 2c.
The network of intra-chain hydrogen bonding in both ligand and receptor adds conformational stabilization to the ligand binding region. This network of hydrogen bonds, especially between Arg-251 and invariant Asp-283
(Kinemage 7 ) of the receptor and between Asn-104 and Tyr-106 of the ligand could serve as a barrier to rotation about the critical Arg-251 residue of the receptor. A restriction of rotation here would facilitate ligand-receptor interaction. It turns out that changing Tyr-106 to a Phe reduces binding affinity 5-fold.
Since there is much sequence variation (primarily as a result of splice variation) in the loop regions of FGF receptor domain D3, the D3 domain of FGF receptors is associated with ligand discrimination. This region is associated with the low affinity loop (defined by the turn between beta loops 8 and 9 of the FGF trefoil structure) of FGF molecules.
There are multiple hydrogen bonding interactions between FGF ligands and the D3 domain of FGF receptors. These form a hydrogen bonding network that is dependent upon the sequence of both ligand and splice variant of the receptor. Many of these H bonds (like ligand Ala-57 to receptor Asp-321) are mediated via a water molecule. The direct hydrogen bond interaction between Glu-96 of FGF 2 and Gln-285 of D3 in
FGFR 2c is particularly important. Ala substitution of this FGF 2 Glu-96 leads to a 1000 fold decrease in binding.
Hydrophobic interactions primarily involve Val-317 of the FGFR 2c interacting with the cavity defined by Tyr-73, Val-88, and Phe-93 of FGF 2. Ala replacement of Val-88 leads to a 10-fold decrease in binding affinity of FGF 2 with
FGFR 2c; Ala replacement of Phe-93 leads to an 80 fold decrease in binding affinity.
The N-terminal regions of FGF are too conformationally mobile to be detected by X-ray experiments. However, when bound to FGF receptors, the N-terminal region is resolvable and appears to participate in ligand-receptor binding. The FGF 2 residue Phe-17 slides into a shallow cavity defined by the D3 region of
FGFR 2c residues Val-280, Pro-286 and Ile-288. A synthetic peptide (residues 13-18) competes with FGF 2 for
FGFR 2c binding. This, and the diversity of N-terminal sequences in FGF species, suggests the flexible N-terminal region may be a significant component in FGF to FGF receptor affiliation.
A summary of all the
critical binding residues is shown in Kinemage 8.
The Kinemages
These kinemages do not render hydrogen atoms, so rendered hydrogen bonds other than the main chain will appear longer than reality since they are drawn between the hetero-atoms involved in the hydrogen bonding.
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1:The FGF 2-FGFR 2c Unit Cell (Dimer of Dimers)
This shows the unit cell as a duplex of the ligand (FGF 2)-receptor (FGFR 2c) dimer complex. The disulphide bond in the receptor IG Domain 2 (D2) between Cys residues 179 and 231 is shown. The sulfate ions in the crystal mark one point of the "basic canyon" region where heparin stabilizes FGF.
View 1 the unit cell duplex of dimers.
View 2 view 1 rotated ninety degrees.
|
77 K |
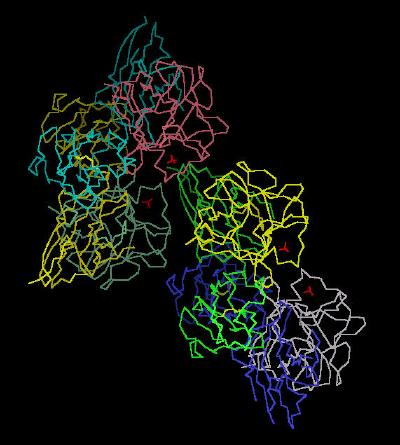 |
| Click on KiNG to see | FGF 2 - FGFR 2c Complex |
Kinemage 2: The FGF 2-FGFR 2c Dimer
Calpha trace of Kinemage 1 with only a single dimer complex present
View 1 the ligand-receptor dimer.
View 2 view 1 rotated ninety degrees.
|
39 K |
 |
| Click on KiNG to see | The FGF 2-FGFR 2c Dimer |
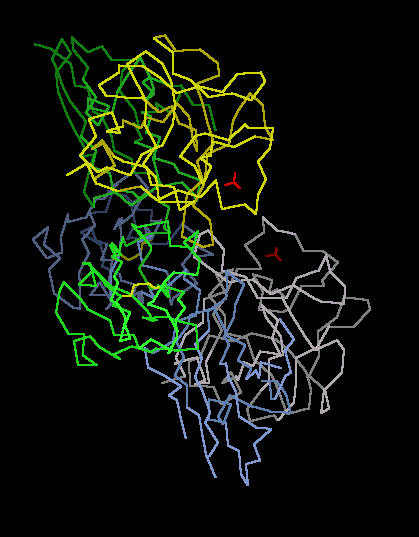
Kinemage 3: A Single FGF 2 Residing in the Ligand Binding Domain (D2 and D3) of FGFR 2c
The Calpha trace of Kinemage 2 with only one FGF 2 to FGFR 2c interaction displayed. This is the parent for all the following kinemages.
View 1 the ligand-receptor dimer.
View 2 view 1 rotated ninety degrees.
|
20 K |
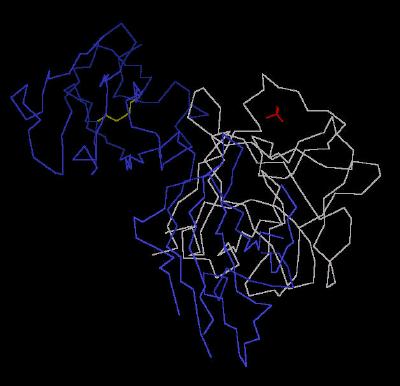 |
| Click on KiNG to see | FGF2 in the Binding Domain of FGFR 2c |
Kinemage 4: A Single FGF 2 Residing in the Ligand Binding Domain (D2 and D3) of FGFR 2c
View 1 the ligand-receptor dimer.
View 2 view 1 rotated ninety degrees.
|
446 K |
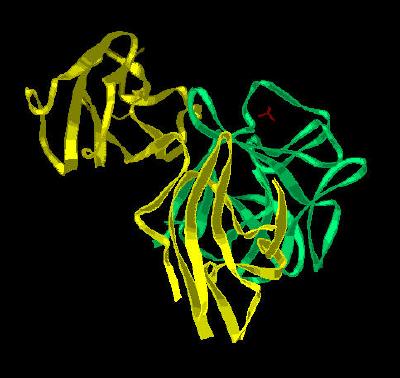 |
| Click on KiNG to see | FGF2 in Ligand Binding Domain of FGFR 2c |
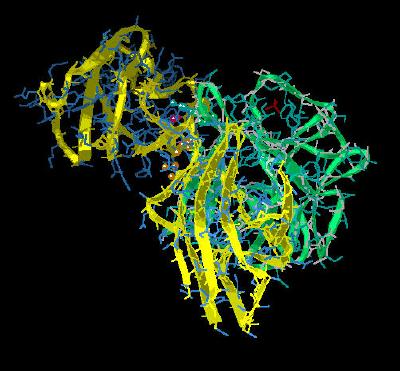
Kinemage 5: Residues Involved in the Binding of FGF 2 to FGFR 2c Domain 2
The high affinity binding region is primarily a site dominated by hydrophobic interactions as the solvent exposed hydrophobic surfaces of both ligand and receptor interact to exclude polar solvent. This is highlighted by FGF residues Tyr-24, Leu-140 and Met-142 packing around Ala-168 of the D2 domain of FGFR 2c. Additionally, Leu-140, Tyr-103 and the aliphatic portion of Asn-102 define a hydrophobic contact volume with Pro-170 of FGFR 2c. Further interactions suggested by the crystal structure include H bonding between the Tyr-24 hydroxyl of FGF 2 and the backbone of Leu-166 and Ala-168 of FGFR 2c. There is also a water mediated hydrogen bond between the hydroxyl of Tyr-103 of FGF 2 and the backbone of Ala-168 of FGFR 2c.
View 1 the ligand-receptor dimer.
View 2 view 1 rotated ninety degrees.
View 3 the hydrophobic packing of residues Tyr-24, Leu-140 and Met-142 of FGF 2 with Ala-168 of the D2 domain of the
FGFR 2c receptor.
View 4 the hydrophobic packing site defined by FGF 2 residues Leu-140, Tyr-103 and the aliphatic portion of Asn-102 surrounding Pro-170 of
FGFR 2c.
View 5 the interaction between Phe-31 of FGF 2 with Leu-166 of FGFR 2c.
View 6 hydrogen bonding sites between ligand (FGF 2) and receptor (FGFR 2c) involving Tyr-24 of FGF 2 hydrogen bonding to the backbone of Leu-166 and Ala-168 of the D2 domain of
FGFR 2c. There is also a water mediated hydrogen bond between Tyr-103 of FGF 2 and Ala-168 of
FGFR 2c.
|
445 K |
 |
| Click on KiNG to see | Residues Binding FGF 2 to FGFR 2c Domain 2 |
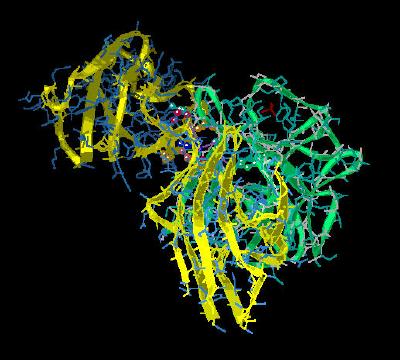
Kinemage 6: Residues Involved in the High Affinity Binding of FGF 2 to the FGFR 2c Linker Region (between D2 and D3)
The hydrogen bonds in this region between the invariant Arg-253 of the FGFR 2c receptor with the backbone of Asn-102 and the side chain of Asn-104 of the FGF 2 ligand are particularly important.
View 1 the ligand-receptor dimer.
View 2 view 1 rotated ninety degrees.
View 3 the hydrophobic interactions of Val-249 and Pro-253 of FGFR 2c with the FGF 2 residues Leu-98 and Pro-141.
View 4 the FGFR 2c Arg-251 interactions with the FGF2 ligand.
View 5 the region where the linker sequence between D2 and D3 of FGFR 2c
joins D2.
Turn on the main chain hydrogen bonds (mc H bonds) and note how the network of main chain hydrogen bonds of the beta sheets serve to restrict rotation of the D3 region with respect to the linker. This restriction of rotation most likely increases the facility of ligand-receptor interaction.
Turn on LL H bond to see the intra-chain FGF 2 hydrogen bond between Asn-104 and Tyr-106.
Turn on RR H bond to see the intra-chain FGFR 2c hydrogen bond between Arg-251 and Asp-283.
|
445 K
|
 |
| Click on KiNG to see | Sites Binding to D2-D3 Linker |
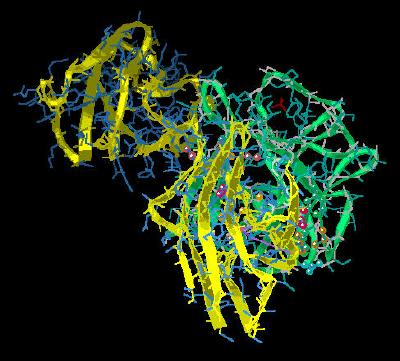
Kinemage 7: Residues Involved in the Low Affinity Binding of FGF 2 to FGFR 2c
This network of hydrogen bonds, especially between Arg-251 and invariant Asp-283
of the receptor and between Asn-104 and Tyr-106 of the ligand could serve as a barrier to rotation about the critical Arg-251 residue of the receptor.
Since there is much sequence variation (primarily as a result of splice variation) in the loop regions of FGF receptor domain D3, the D3 domain of FGF receptors is associated with ligand discrimination. This region is associated with the low affinity loop (defined by the turn between beta loops 8 and 9 of the FGF trefoil structure) of FGF molecules.
There are multiple hydrogen bonding interactions between FGF ligands and the D3 domain of FGF receptors. These form a hydrogen bonding network that is dependent upon the sequence of both ligand and splice variant of the receptor. Many of these H bonds (like ligand Ala-57 to receptor Asp-321) are mediated via a water molecule. The direct hydrogen bond interaction between Glu-96 of FGF 2 and Gln-285 of D3 in
FGFR 2c is particularly important. Ala substitution of this FGF 2 Glu-96 leads to a 1000 fold decrease in binding.
Hydrophobic interactions primarily involve Val-317 of the FGFR 2c interacting with the cavity defined by Tyr-73, Val-88, and Phe-93 of FGF 2. Ala replacement of Val-88 leads to a 10-fold decrease in binding affinity of FGF 2 with
FGFR 2c; Ala replacement of Phe-93 leads to an 80 fold decrease in binding affinity.
View 1 the ligand-receptor dimer.
View 2 view 1 rotated ninety degrees.
For views 3 & 4, turn on the main chain to see H bonds from residues to backbone.
Also, turning on water visualizes the H bond between ligand Ala-57 (backbone) and receptor Asp-321 via an ordered water molecule
View 3 the FGF2 region that interacts with D2 of FGFR 2c.
View 3 the hydrogen bonding between FGF 2 and FGFR 2c at FGF 2 residue Gln-56.
View 4 the hydrogen bonding between FGF 2 and FGFR 2c centering around FGF residue Asn-104.
View 5 the hydrophobic residues involved in the ligand-receptor
interaction: Val-317 of the
FGFR 2c receptor interacting with FGF 2 residues Tyr-73, Val-88 and Phe-93.
View 6 the N terminal region of FGF 2 centered in the vicinity of residue Phe-17 which fits into a shallow hydrophobic valley defined by
FGFR 2c residues Val-280, Pro-286, and Ile-288.
|
447 K |
 |
| Click on KiNG to see | FGF Low Affinity Binding Sites |
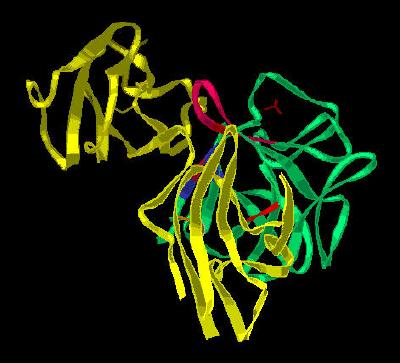
Kinemage 8: Ribbon Representation of the Critical Residues Involved in the FGF 2 Interaction with FGFR 2c
View 1 the unit cell duplex of dimers.
View 2 view 1 rotated ninety degrees.
View 3 the FGF2 region that interacts with D2 of FGFR 2c.
View 4 the FGF2 region that interacts with the D2-D3 Linker region of
FGFR 2c.
View 5 the FGF2 region that interacts with D3 of FGFR 2c.
View 6 the ordered N-terminal region of FGF 2.
|
461 K |
 |
| Click on KiNG to see | Ribbon Rendering Of Binding Sites |
The Sequences
FGF Chain A
GHFKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYL
AMKEDGRLLASKSVTDECFFFERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQ
KAILFLPMSAKS
FGF Chain B
GHFKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYL
AMKEDGRLLASKSVTDECFFFERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQ
KAILFLPMSAKS
FGF Chain C
GHFKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYL
AMKEDGRLLASKSVTDECFFFERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQ
KAILFLPMSAKS
FGF Chain D
GHFKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYL
AMKEDGRLLASKSVTDECFFFERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQ
KAILFLPMSAKS
FGFR Chain E
NSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGG
YKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANA
STVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEV
LYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGRE
FGFR Chain F
NSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGG
YKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANA
STVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEV
LYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGRE
FGFR Chain G
NSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGG
YKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANA
STVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEV
LYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGRE
FGFR Chain H
NSNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGG
YKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANA
STVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEV
LYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGRE
For Chain A (X-Ray resolved residues are 17-145 of the human FGF sequence)
Unresolved N-Terminal: GH
X-Ray Resolved: FKDPKRLYCKNGGFFLRIHPDGRVDGVREKSDPHIKLQLQAEERGVVSIKGVSANRYL
AMKEDGRLLASKSVTDECFFFERLESNNYNTYRSRKYTSWYVALKRTGQYKLGSKTGPGQKAILFLPMSAK
Unresolved C-Terminal : S
For Chain E (X-Ray resolved residues are 150-359 of the human FGFR 2c sequence)
Unresolved N-Terminal: NSNN
X-ray resolved: KRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGG
YKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANA
STVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEV
LYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVL
Unresolved C-Terminal: PAPGRE
Source:
Human sequence; The structural coordinates were taken from the Brookhaven Database file1EV2.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.