 |
 |
|
Stick View of Su-5402 |
Inhibitor Surface Within Tyrosine Kinase Domain |
|
Inhibitor SU-5402 Bound to the Tyrosine Kinase Domain of FGFR 1 by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
Inhibitor SU-5402 Bound to the Tyrosine Kinase Domain of FGFR 1
FGF molecules function by interacting with FGF receptors. These receptors typically contain Ig-like binding domains (with domains D2 and D3 being most involved in FGF-FGF receptor interactions), a short transmembrane spanning domain and a cytoplasmic component that possesses tyrosine kinase activity. An essential component of biological activity is the heparin-mediated dimerization of the FGF receptor.
Most FGF receptors contain a tyrosine kinase motif downstream from the ligand binding domain and membrane spanning region.
Most FGF receptors contain a tyrosine kinase motif downstream from the ligand binding domain and membrane spanning region.
Since FGF is implicated in a variety of growth disorders and cancers, it seems
reasonable that blocking the FGF signal activity via inhibition of the tyrosine
activity would be of therapeutic value. So, there is currently a search for good
inhibitors of this system.
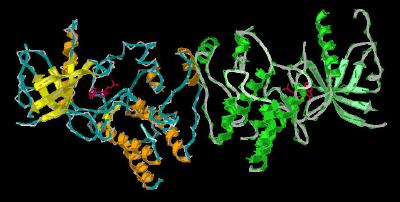
This crystal structure
for 1FGI is from an engineered protein that represents residues 404-761 (sequence below) of the human FGF receptor 1 (FGFR 1). The X-ray structure of the unit cell dimer
(Kinemage 1 and Kinemage 2 ) shows that the inhibitor SU-5402 {3-[(3-(2-carboxyethyl)-4-methylpyrrol-2-yl)methylene]-2-indolinone}
resides in
a cleft between the two distinct lobes of the tyrosine kinase domain of the receptor, with the oxy-indole ring near the volume occupied by the adenine base of ATP. (This is easiest to visualize in
Kinemage 3). The characteristics of the unit cell for this
structure are summarized at pdbsum.
The SU-5402 inhibitor resides in a cleft of the tyrosine binding region of FGFR 1 that is defined by hydrophobic residues Leu-484, Phe-489, Val-492, Ile-545, Val-561, Tyr-563, Ala-254, and Gly-567. The position of the SU-5402 inhibitor within the binding cleft is anchored by hydrogen bonding between the inhibitor oxyindole ring N1 and the backbone carbonyl of Glu-562, the backbone amide of Ala-564 to the oxyindole ring carbonyl, and the side chain amide of Asn-568 to oxygen of the inhibitor carboxyethyl group.
Of particular significance is the oxy-aromatic interactions of the hydrogen atoms of the plane of Phe-489. All five phenyl group hydrogens are interacting with near-by oxygen
atoms: each of the two oxygen atoms of the inhibitor carboxyethyl group, the backbone carbonyl of Arg-627 and the side chain
oxygen atoms of Asn-628 and Asp-641. This network of oxy-aromatic interactions and hydrogen bonds most likely explains the preference of SU-5402
for the FGFR 1 receptor. Also, the area of this receptor's "phenyl loop" is disordered with other inhibitors, but quite well resolved with
SU-5402, These interactions are highlighted in
Kinemage 4.
The SYBYL stick rendering of SU-5402 and the surface contour of
this compound within the binding cleft of the Tyrosine Kinase Domain of FGFR 1
is shown below:
 |
 |
|
Stick View of Su-5402 |
Inhibitor Surface Within Tyrosine Kinase Domain |
|
Atom |
Color |
Structure |
Color |
|
|
Carbon |
white |
Peptide alpha |
magenta |
|
|
Hydrogen |
cyan |
Peptide beta | yellow | |
|
Oxygen |
red |
Peptide coil | cyan | |
|
Nitrogen |
blue |
Inhibitor Surface |
green dots |
The Kinemages:
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1: The Crystal Dimer FGFR 1 Bound To the Inhibitor SU-5402
View 1 The Dimer
View 2 Binding Cleft, Chain A from "front"
View 3 Binding Cleft, Chain A from "top"
View 4 Binding Cleft, Chain B from "front"
View 5 Binding Cleft, Chain B from "top"
|
41 K |
 |
| Click on KiNG to see | The Unit Cell |
Kinemage 2: The Crystal Dimer FGFR 1 Bound To the Inhibitor SU-5402
This kinemage adds main chain, side chains, main chain hydrogen bonds, ribbon and cartoon rendering to kinemage 1.
View 1 The Dimer
View 2 Binding Cleft, Chain A from "front"
View 3 Binding Cleft, Chain A from "top"
View 4 Binding Cleft, Chain B from "front"
View 5 Binding Cleft, Chain B from "top"
|
1.04 M |
 |
| Click on KiNG to see | Cartoon Rendering |
Kinemage 3: Inhibitor SU-5402 Bound To FGFR 1 Tyrosine Kinase Domain
View 1 The monomer
View 2 Binding Cleft, "front"
View 3 Binding Cleft, "top"
|
538 K |
 |
| Click on KiNG to see | Monomer of the Inhibitor-Receptor Complex |
Kinemage 4: Inhibitor SU-5402 Bound To FGFR 1 Tyrosine Kinase Domain Interactions
The SU-5402 inhibitor resides in a cleft defined by hydrophobic residues Leu-484, Phe-489, Val-492, Ile-545, Val-561, Tyr-563, Ala-564, and Gly-567. The position of the SU-5402 inhibitor within the binding cleft is anchored by hydrogen bonding between the inhibitor oxyindole ring N1 and the backbone carbonyl of Glu-562, the backbone amide of Ala-564 to the oxyindole ring carbonyl, and the side chain amide of Asn-628 to oxygen of the inhibitor carboxyethyl group. All five phenyl group hydrogens are interacting with near-by oxygen atoms: each of the two oxygen atoms of the inhibitor carboxyethyl group, the backbone carbonyl of Arg-627 and the side chain oxygen atoms of Asn-628 and Asp-641.
View 1 The Monomer
View 2 Binding Cleft, "front"
View 3 Binding Cleft, "top"
View 4 H Bonding Interactions
View 5 Oxy-aromatic interactions
|
537 K |
 |
| Click on KiNG to see | Inhibitor-Receptor Interactions |
Sequence: The X-ray resolved residues span residues 464-762 of the human FGFR 1 sequence
Unresolved N-terminal: MVAGVSEY
X-ray resolved: ELPEDPRWELPRDRLVLGKPLGEGAFGQVVLAEAIGLDKDKPNRVTKVAVKM
LKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEY
SYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHI
DYYKKTTNGRLPVKWMAPEALFDRIYTHQSDVWSFGVLLWEIFTLGGSPYPGVPVEELFKLLKEGHRM
DKPSNCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRIVALTS
Unresolved C-terminal: NQE
Source:
Residues 464-762 of human FGFR 1 was isolated from transfected Spodoptera frugiperda insect cells; structural coordinates were taken from the Brookhaven Database file 1FGI.
The engineered sequence contained three residues that were different from human:
Cys-488 and 584 were changed to Ser to prevent disulfide oligiomerization.
Leu-457 was changed to Val to create an aNcol cloning site.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.