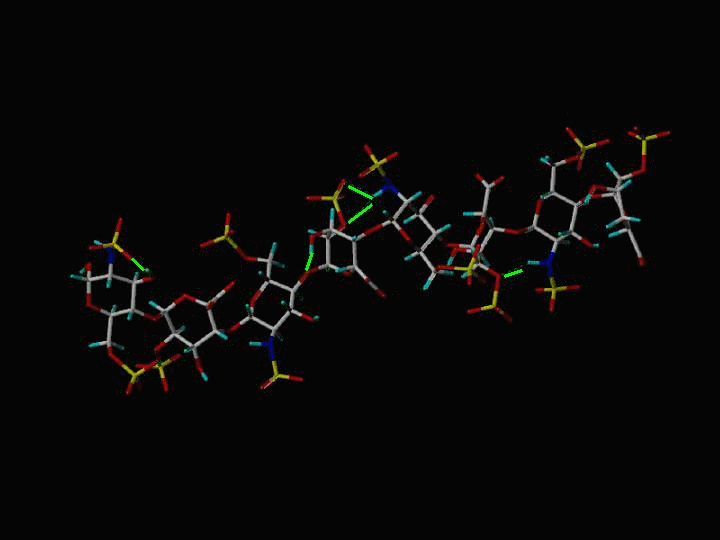
|
by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to:
Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
Heparin
Heparins are found in a variety of cells and tissues interacting with an abundance of physiological effecter proteins including morphogens, growth factors, and enzymes. Heparins are complex acidic polysaccharides that are characterized by a disaccharide repeat unit of a-D-glucosamine 1-4 linked to uronic (a-L-iduronic / b-D-glucuronic) acid. Structural heterogeneity primarily arises from the number of polysaccharide repeats present and the chemical nature of substitution (acetylation or sulfation) of the sugar monomers. The sites of sulfation include the 2-O position of the uronic acid and the N, 3-O, and 6-O positions of the glucosamine. The abundant negative charges from sulfation at multiple sites of the sugar polymer create an overall acidic nature for the heparin polymer. In addition, the N position of the glucosamine can be acetylated.
Heparins adopt a helical structure with local geometry governed by hydrogen bond formation, the nature and extent of the chemical modifications, and the conformation (chair, boat or skewed) of each sugar monomer.
Kinemage 1 is based on the heparin extracted from the crystal structure of the FGF 2-FGFR 1-Heparin (2:2:2) complex.
The Heparin decasaccharide shown below is modeled as a helix of repeating disaccharide units of a-D-glucosamine (GlcN) and a-L-iduronic acid (IdoA) joined by a-1-4 linkages. Each disaccharide unit has three sulfates: one at the 2-hydroxyl group of IdoA (forming 1,4-dideoxy-O2-sulfo-glucuronic acid ... IDU) and two at the 2-amino and 6-hydroxyl groups of GlcN (forming N,O, 6-disulfo-glucosamine ... SGN). The helix conformation is stabilized by internal hydrogen bonding.
The Stick Rendering of Heparin

|
Atom |
Color |
|
C |
White |
|
H |
Cyan |
|
N |
Blue |
|
O |
Red |
|
S |
Yellow |
| H-Bonds | Green Lines |
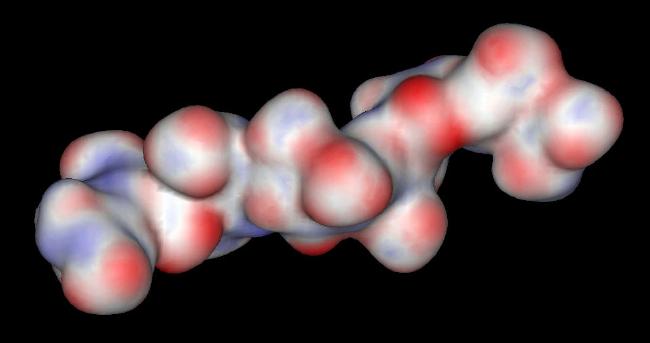
The sulfate and carboxylate groups form a negative charge potential that lines the ridge of the Heparin helix such that a negative center is found every 17-19 angstroms. Heparin polysaccharides are quite polar entities with a non-reducing end (O4) and a reducing end (O1). In the crystal structure of the FGF-FGFR-Heparin (2:2:2) dimer complex, the decasaccharides bind with their non-reducing ends (1,4-dideoxy-5-dehydro-O2-sulfo-glucuronic acid ... UAP) in the center of the FGFR "basic canyon" and continue outward from the interior of the complex onto the high-affinity heparin binding sites of the FGF ligands. This maintains the two-fold symmetry of the dimer complex. (Note that a single, longer species of Heparin would not maintain the 2-fold symmetry of the complex). The potential surface of heparin is shown below. The negative centers are red.
The Potential Surface of Heparin
\
The glucosamine rings of SGN are all found in a chair conformation. The rings of the other sugars (UAP and IDU) are in either a chair or a skewed boat conformation. This suggests that glucuronic acid ring can adopt multiple orientations depending on the specific FGF or FGFR contact points. This conformational flexibility of the glucuronic acid rings most likely plays a role in specific ligand and/or receptor recognition.
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1: Heparin
The Heparin decasaccharide shown below is modeled as a helix of repeating disaccharide units of a-D-glucosamine (GlcN) and a-L-iduronic acid (IdoA) joined by a-1-4 linkages. Each disaccharide unit has three sulfates: one at the 2-hydroxyl group of IdoA (forming 1,4-dideoxy-O2-sulfo-glucuronic acid ... IDU) and two at the 2-amino and 6-hydroxyl groups of GlcN (forming N,O, 6-disulfo-glucosamine ... SGN).
View 1 Heparin
View 2 intra-molecular H-bond 302-303
View 3 intra-molecular H-bond 304-305 (to both sugar O and sulfate O)
View 4 intra-molecular H-bond 305-306
View 5 intra-molecular H-bond 308-308
Each carbohydrate ring has both a substructure number assigned by the PDB structural coordinate file and an alphabetic identifier. This correspondence is shown below:
| Sub-Structure | Designation | Sub-Structure |
Designation |
|
| 301 | A | 305 | E | |
| 302 | B | 306 | F | |
| 303 | C | 307 | G | |
| 304 | D | 308 | H |
|
16 K |
 |
| Click On KiNG to see |
Heparin |
Source:
The heparin molecule was extracted (using SYBYL) from the Brookhaven Database file 1FQ9 (Chain E). The intra-molecular hydrogen bonds were assigned using the View Static H Bonds option of SYBYL. The potential surface was generated using the Windows version of AccelyrSys Viewer Lite.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular &
Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.