|
FGF 1, 4, 7, 8b, 9, 10, 12 & 19 Superimposed Upon FGF 2 by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
FGF Structural Comparisons
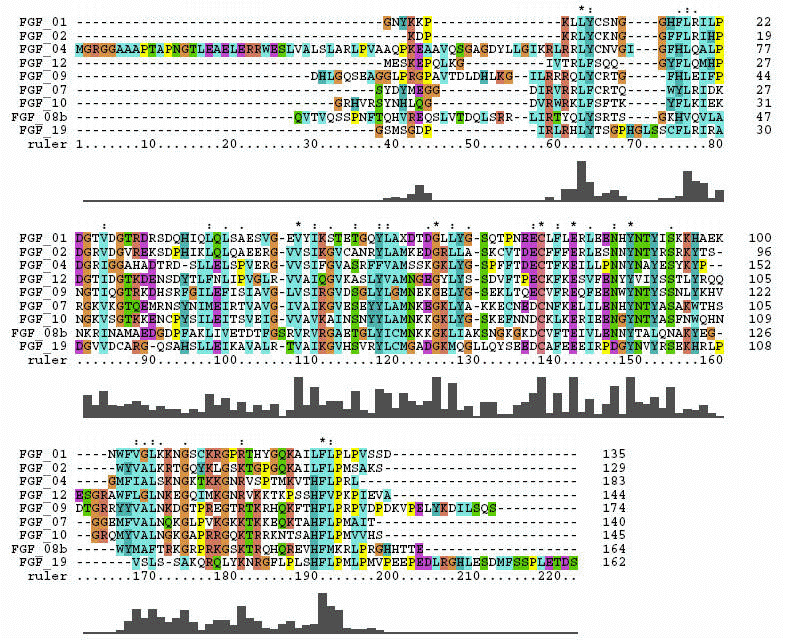
FGF molecules share a similar 3-Dimensional architecture (beta trefoil motif). Within this basic overall 3-D shape, there are structural differences that can rationalize different binding affinities for their FGF receptors with subsequent variation in biological activity. This page highlights two tools used to examine protein structural comparisons: sequence alignments to spot regions of identical or similar amino acid composition and a structural comparison kinemage to visualize sequence-dependant local structural variations.
The FGF crystal structure sequences that were used in the structural overlay were aligned using Clustal X. This alignment is shown below:

Sequence comparisons can furnish insights into conserved or variable portions of a molecular species, predict some secondary structure characteristics and suggest protein motifs common to known proteins. These tools can provide clues as to areas of the molecule that are biologically interesting. Real molecules, however, do not have gaps in their sequences, so, whenever possible, comparison of 3-D architecture is used to give a better "picture" of molecular behavior. After all, the basic theology of organic chemistry is that form and function are intimately related.
A single click on the KiNG logo will launch the kinemage. The number under the KiNG logo is the download size (in bytes).
Use the left mouse button to rotate the molecule.
If the KiNG logo is absent or the application does not launch, see Gray Box Error.
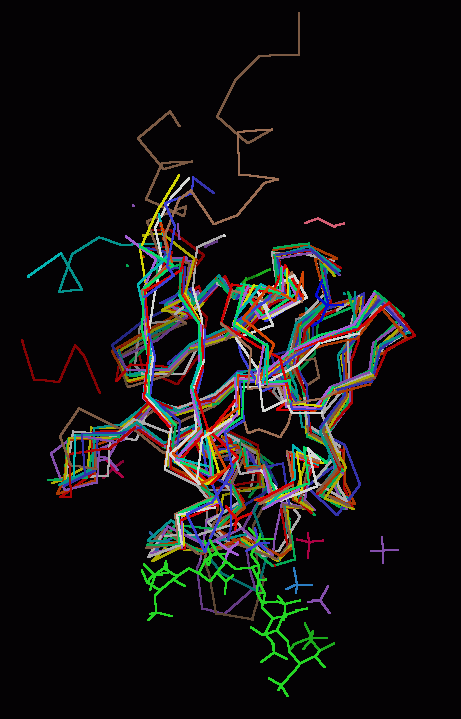
FGF 1, 4, 7, 8b, 9, 10, 12 & 19 Superimposed on FGF 2
The backbones of
human FGF 1, 4, 7,
8b, 9, 10, 12, and 19 were
superimposed upon the backbone
of FGF 2. Basic FGF (FGF 2) was chosen as the template or
reference structure because it universally binds to all the
known FGF receptors and the complete core sequence is well-resolved by X-ray
diffraction.
Other FGF molecules were individually superimposed on
the FGF 2 template using a single pass of the "magic fit" application of
molecular visualization package Deep View. The intent was not to develop the
best possible RMS template, but to quickly (using commonly available freeware software)
create an image that highlighted the areas of structural divergence among the
various FGF
molecules, since it is the areas of divergence that most likely account for
differences in observed biological activity.
Each FGF molecule is
represented as a Calpha backbone trace, with side chains, disulphide bonds, and
ribbon rendering available. In addition, non-water associated hetero-atoms from the crystal unit
cell for each FGF molecule and heparin are shown for purposes of
visual orientation.
Finally, the three distinct folds of the beta trefold motif are indicated by ribbon segments. The segments are numbered according to the FGF 2 template sequence.
The Beta Sheets Of the Trefoil Motif
|
Fold 1 |
Fold 2 |
Fold 3 |
| Sheet | Residues | Sheet | Residues | Sheet | Residues |
| 1 | 28-34 | 4 | 60-67 | 8 | 101-107 |
| 2 | 37-4 | 5 | 59-76 | 9 | 110-117 |
| 3 | 46-51 | 6 | 79-85 | 10 | 122-127 |
| 12 | 146-152 | 7 | 89-93 | 11 | 130-133 |
The Kinemage
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
View 1
the FGF backbones
View 2 the low affinity (discrimination) loop.
View 3 the heparin binding region (the "basic Canyon")
View 4 the N & C terminal junction
|
1.52 M |
 |
| Click On KiNG to see |
FGF Backbones |
The FGF protein family constitutes a superb example of conserving a basic three-dimensional geometry with sequence variation to modify biological activity.
Coordinates Source:
Structural Coordinates for the FGF species were taken from Brookhaven Database files 1AXM (FGF1 & heparin), 1BFF (FGF2), 1IJT (FGF 4), 1QQK (FGF 7), 1IHK (FGF 9), 1Q1U (FGF 12), and 1PWA (FGF 19). The structures for FGF 8b (2FDB) and FGF 10 (1NUN)) were extracted with SYBYL from coordinates for a ligand-receptor complex.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular &
Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.