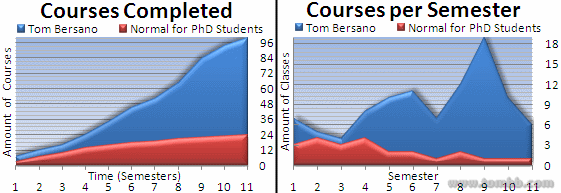
- Everything (Compact)
- Academics
- Teaching
- Research
- Publications
- Labs & Industry - Timeline
- Everything (Printable)
 Post-Doctoral Training in(Completed in '05, '08, '10)
Post-Doctoral Training in(Completed in '05, '08, '10)
MED, EECS, BME, PHYSInt.Medicine/Oncology:'10-'11Cancer Cell Capture Technologies.Electrical Engineering:'08-'10MEMS for Single-Cell Biology.Biomed. Engineering:'05-'08Microfluidic Automation.Physics:'04-'05BioInformatics, Quantum Computing.
 Doctorate (Ph.D.) in(Graduated Dec 2003)
Doctorate (Ph.D.) in(Graduated Dec 2003)
Computer Engineering - AIMy PhD Dissertation was:"Computer evolution of gene circuits for cell-embedded computation, biotechnology, and as a model for evolutionary computation". Links to abstract [A], table of contents [T], ProQuest[P]My PhD Advisor was:John H. Holland. He authored several books in Artificial Intelligence and Complex Systems in which he established widely used optimization and AI techniques such as Genetic Algorithms (computer evolution of designs and solutions). He was awarded the prestigious MacArthur Genius Grant
 Master (M.S.) in(Graduated Dec 2001)
Master (M.S.) in(Graduated Dec 2001)
BioChemistry
 Master (M.S.) in(Graduated May 2001)
Master (M.S.) in(Graduated May 2001)
Kinesiology
 Master (M.S.E.) in(Graduated Dec 2001)
Master (M.S.E.) in(Graduated Dec 2001)
BioMedical Engineering
 Master (M.Eng.) in(Graduated May 1999)
Master (M.Eng.) in(Graduated May 1999)
Aerospace Engineering
 Cert. of Grad Studies in(Graduated May 1999)
Cert. of Grad Studies in(Graduated May 1999)
Complex Systems
 Master (M.S.E.) in(Graduated May 1997)
Master (M.S.E.) in(Graduated May 1997)
Computer Engineering
 Master (M.S.) in(needed just 3 more term papers)
Master (M.S.) in(needed just 3 more term papers)
Philosophy
 Master (M.S.E) in(needed just 3 more classes)
Master (M.S.E) in(needed just 3 more classes)
Elec.Eng. - Circuits & BioSys.
 Master (M.S) in(needed just 1 more project report)
Master (M.S) in(needed just 1 more project report)
Psychology - Cog. Neurosci.
 Bachelor (B.S.E) in(Graduated with Honors May 1996)
Bachelor (B.S.E) in(Graduated with Honors May 1996)
Computer Engineering

 Undergraduate Honors in(Dean's List 1995-1996)
Undergraduate Honors in(Dean's List 1995-1996)
Computer Engineering
Specifically, I believe that good teaching requires communicating excitement about the subject and a deep understanding that goes beyond stating a list of facts and rather tries to tie them into interesting and memorable stories and intuitive analogies. ...
 2001:Logic Design EECS270
2001:Logic Design EECS270
My UofM Ranking as Teacher:95% 2001: Intro to Logic Design (EECS270)Course Description: Introduction to Logic Design Binary and non-binary systems, Boolean algebra digital design techniques, logic gates, logic minimization, standard combinational circuits, sequential circuits, flip-flops, synthesis of synchronous sequential circuits, PLA's, ROM's, RAM's, arithmetic circuits, computer-aided design. Laboratory includes hardware design and CAD experiments.My Ranking as a Teacher in this course, from the University of Michigan's Official Student Evaluations96%Was available throughout the designated lab hours.98%Thoroughly understood the subject matter.93%Was sensitive to the level of student comprehension.91%Explained the material clearly and understandably.96%Had no English language problem.96%Overall, the instructor was effective.What my students said about me in official evaluations:
2001: Intro to Logic Design (EECS270)Course Description: Introduction to Logic Design Binary and non-binary systems, Boolean algebra digital design techniques, logic gates, logic minimization, standard combinational circuits, sequential circuits, flip-flops, synthesis of synchronous sequential circuits, PLA's, ROM's, RAM's, arithmetic circuits, computer-aided design. Laboratory includes hardware design and CAD experiments.My Ranking as a Teacher in this course, from the University of Michigan's Official Student Evaluations96%Was available throughout the designated lab hours.98%Thoroughly understood the subject matter.93%Was sensitive to the level of student comprehension.91%Explained the material clearly and understandably.96%Had no English language problem.96%Overall, the instructor was effective.What my students said about me in official evaluations:I really enjoyed being in Tom's lab section this year. He was very inspiring and knowledgeable!
Tom did a great job in an extremely frustrating class.
Tom was a great GSI.
 2001:C++ Progr. EECS280
2001:C++ Progr. EECS280
My UofM Ranking as Teacher:94% 2001: Intro to C++ Programming (EECS280)Course Description: Techniques and algorithm development and effective programming, top-down analysis, structured programming, testing, and program correctness. Program language syntax and static and runtime semantics. Scope, procedure instantiation, recursion, abstract data types, and parameter passing methods. Structured data types, pointers, linked data structures, stacks, queues, arrays, records, and trees.My Ranking as a Teacher in this course, from the University of Michigan's Official Student Evaluations97%Was available throughout the designated lab hours.97%Thoroughly understood the subject matter.96%Was sensitive to the level of student comprehension.89%Explained the material clearly and understandably.95%Had no English language problem.92%Overall, the instructor was effective.What my students said about me in official evaluations:
2001: Intro to C++ Programming (EECS280)Course Description: Techniques and algorithm development and effective programming, top-down analysis, structured programming, testing, and program correctness. Program language syntax and static and runtime semantics. Scope, procedure instantiation, recursion, abstract data types, and parameter passing methods. Structured data types, pointers, linked data structures, stacks, queues, arrays, records, and trees.My Ranking as a Teacher in this course, from the University of Michigan's Official Student Evaluations97%Was available throughout the designated lab hours.97%Thoroughly understood the subject matter.96%Was sensitive to the level of student comprehension.89%Explained the material clearly and understandably.95%Had no English language problem.92%Overall, the instructor was effective.What my students said about me in official evaluations:I really enjoyed this discussion section! I would never have survived 280 without a discussion leader who totally knew his subject material and who was funny, approachable, and overall excellent.
Very competent of subject matter - could always answer questions. Has a very good knack for presenting analogies that demonstrate concepts clearly (and often humorously!)
I found your teaching style useful, informative, and refreshing...
I liked Tom, he knew the material well.
 1997:Computer Arch. CS370
1997:Computer Arch. CS370
My UofM Ranking as Teacher:N/A 1997: Computer Organization Architecture and Design (EECS370)Course Description: Basic concepts of computer organization and hardware. Instructions executed by a processor and how to use these instructions in simple assembly-language programs. Stored-program concept. Data-path and control for multiple implementations of a processor. Performance evaluation, pipelining, caches, virtual memory, input/output.What my students said about me in official evaluations:
1997: Computer Organization Architecture and Design (EECS370)Course Description: Basic concepts of computer organization and hardware. Instructions executed by a processor and how to use these instructions in simple assembly-language programs. Stored-program concept. Data-path and control for multiple implementations of a processor. Performance evaluation, pipelining, caches, virtual memory, input/output.What my students said about me in official evaluations:Tom should receive an award for the work he put into this class. He assumed most of the responsibility for the class after all.
Tom appeared to have a thorough knowledge of the material.
- BioTechniques Journal (2009)
- IEEE Transactions on Evolutionary Computation (1999, 2000, 2003)
- Bio-Techno Conference (2010, 2011, 2012)
- Genetic and Evolutionary Computation Conference (2002, 2003, 2004, 2005, 2006, 2007, 2008, 2009, 2010, 2011)
- Evolutionary Computation (2000)
- Genetic Programming Conference(1997-1998)
- 2010Bio+Micro
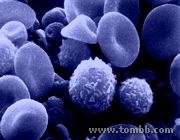 Circulating Tumor Cells
Circulating Tumor Cells- Fig1
 Figure 1: An SEM image of blood1, showing normal circulating cells that need to be separated and distinguished from metastatic cancer cells.
Figure 1: An SEM image of blood1, showing normal circulating cells that need to be separated and distinguished from metastatic cancer cells. - Fig2
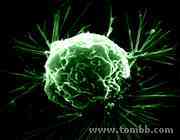 Figure 2: An SEM image of a cancer cell2, anchoring itself on a substrate.
Figure 2: An SEM image of a cancer cell2, anchoring itself on a substrate. - Fig3
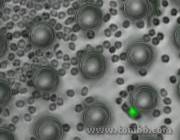 Figure 3: A zoomed-out view of a Circulating Tumor Cell (green) in one of my devices, about to be separated from human blood cells (gray).
Figure 3: A zoomed-out view of a Circulating Tumor Cell (green) in one of my devices, about to be separated from human blood cells (gray). - Video1
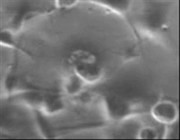 Video 1: Since this project is still ongoing, this video just shows some unrelated data I collected on cancer cells rather than the final device.
Video 1: Since this project is still ongoing, this video just shows some unrelated data I collected on cancer cells rather than the final device.
Separating Circulating Tumor Cells from Blood.The Problem: Metastasis is a dangerous stage of cancer progression in which cancer cells start to shed into the bloodstream and can eventually form multiple secondary tumors. The number of these Circulating Tumor Cells (CTC) in a patient can be used successfully to predict cancer outcome and there are already a few commercial and clinical systems to do this. If we could isolate these cells we could test what treatments might be most effective against them but attempts to do this have not yet been successful. In addition, the existing systems are large and expensive and rely on antibodies or immunomagnetic particles against very specific types of cancer cells. Thus they are not able to detect important cells like cancer stem cells that do not express those antigens, and they only work with fixed (dead) cells, which makes it impossible to do useful tests like drug-resistance on these cells.
To address these problems, I developed a new technology that can separate and capture circulating tumor cells from blood in real time and without relying on antibodies or magnetic particles or fixation techniques, leaving cells alive for further tests, and can be fabricated in microchip size and inexpensive materials. We are currently testing this technology with human blood and cancer cells and with cancer patients' blood.- Fig1
- 2009Bio+Micro
 Cancer Stem Cells
Cancer Stem Cells- Fig1
 Figure 1: Isolated cancer cells (green) in our device, enabling a new level of drug discovery.
Figure 1: Isolated cancer cells (green) in our device, enabling a new level of drug discovery. - Fig2
 Figure 2: After several days, differences in growth speed between microchambers reveal cell subtypes and drug resistance.
Figure 2: After several days, differences in growth speed between microchambers reveal cell subtypes and drug resistance. - Fig3
 Figure 3: A close-up of our microfluidic device, hundreds of microchambers in just a few millimeters in size.
Figure 3: A close-up of our microfluidic device, hundreds of microchambers in just a few millimeters in size. - Video1
 Video 1: Cells are hydrodynamically separated and captured, one clone per chamber.
Video 1: Cells are hydrodynamically separated and captured, one clone per chamber.
Project Year: 2009-2010.
Location: Univ.of Michigan, USA.
Collaborators:Integrated Micro-systems Lab and UM Cancer Center.Single-Cell Device to Identify Cancer Stem CellsThe Problem: Recent evidence in Cancer research strongly indicates that cancers often consist of a mixture of normal cancer cells and a low number of special and more drug-resistant "cancer stem cells". When isolated and reimplanted, cancer stem cells are much more likely to form new tumors and cause relapses.
This means that most of our cancer therapeutics were selected by the wrong criteria, looking at the total average number of cancer cells killed, while the difference between a drug that killed none of the cancer stem cells and one that killed all of them could be lost in the average.
Another problem is that we can currently only identify cancer stem cells in a few specific types of cancers and lack the markers and tools to identify them in a more general way.
To address these problems, we devised a microassay chip that allows us to separate a population of cancer cells into arrays of isolated single cells, identify which of them might be a cancer stem cell by its speed and ability to grow, and then test different drug treatments on these identified subgroups, which might help us find drug combinations that kill both the cancer stem cells and the normal cancer cells.- Fig1
- 2009Bio+Micro
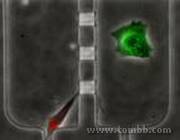 Cells Interactions
Cells Interactions- Fig1
 Figure 1: A cancer cell (green) and muscle cell interact chemically without direct contact on chip (amplified by close positioning and micro scale).
Figure 1: A cancer cell (green) and muscle cell interact chemically without direct contact on chip (amplified by close positioning and micro scale). - Fig2
 Figure 2: We screen for both contact and non-contact interactions through cell pairs in various arrangements across hundreds of microchambers.
Figure 2: We screen for both contact and non-contact interactions through cell pairs in various arrangements across hundreds of microchambers. - Fig3
 Figure 3:Single cells from different tissue types are captured one at a time and precisely placed in direct contact of each other.
Figure 3:Single cells from different tissue types are captured one at a time and precisely placed in direct contact of each other.
Project Year: 2009-2010.
Location: Univ.of Michigan, USA.
Collaborators:Integrated Micro-systems Lab and UM Cancer Center.Observing Interactions Between Specific Cell PairsThe Problem:Cells in our body communicate and organize themselvesboth by contact interactions and by chemical signals. For example, when a salamander has to regrow its tail, its stem cells will coordinate which of them regrows which part. Another example of this is cancer, which often will communicate with the surrounding heathy cells and cause them to grow new blood vessels to feed the tumor. But studying these signals with conventional methods is difficult as it relies on averaged measurement of millions of cells, which hides imortant details and makes high-throughput testing problematic.
To address these problems, we proposed a new microfluidic device which can analyze pairwise cell interactions in isolated microchamber arrays, enabling hundreds of simultaneous tests and potential high-throughput screening of useful cell pairings. Soluble factors secreted by cells in each pair can quickly reach high physiological concentrations when trapped in a small chamber in proximity to each other, rather than being diluted in larger amounts of media as in conventional methods. Media exchange for cell viability is achieved by quick flow-through washes and media perfusion alternated with media isolation phases to expose cells to accumulated signals.- Fig1
- 2008AI+Bio+Micro
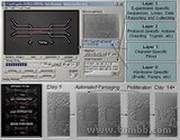 Automated Cell Biology.
Automated Cell Biology.-
- Video1
 Video 1: Cells rearranging over several days under constant observation and guidance of my automatic cell biology system.
Video 1: Cells rearranging over several days under constant observation and guidance of my automatic cell biology system. - Video2
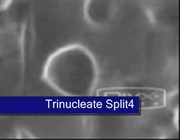 Video 2: My automated system makes it possible to observe rare events such as cells dividing 3way instead of the conventional 2way.
Video 2: My automated system makes it possible to observe rare events such as cells dividing 3way instead of the conventional 2way. - Fig1
 Figure 1: Cells growing, dividing and crawling after weeks of computer-controlled experiments inside my automated microchips.
Figure 1: Cells growing, dividing and crawling after weeks of computer-controlled experiments inside my automated microchips.
Fully-Automated Cell BiologyThe Problem: While biotechnologies like genome sequencing and combinatorial chemistry have benefited from great advances in automation, molecular cell biology experiments involving mammalian cell culture still require continuous manual operation and ad-hoc setup by human researchers. If for example scientists wanted to look at the effect of an anti-aging treatment on cells, they would need to be prepared to carefully handle and maintain cells every other day for several months.
To address these problems, I designed, fabricated and developed a new type of fully automated microfluidic device. Through remote communication these fully-programmable systems can perform long-term cell-passaging outside of incubators, allowing real-time continuous monitoring and tracking of individual cells. My system can also be controlled remotely to make changes to the experimental script in real time, and it sends me email reports at programmed intervals complete with cell microscopy images and video updates. - Video1
-
- 2010Micro
 Fluid Micro-Circuits
Fluid Micro-Circuits- Fig1
 Figure 1: A microscale fluidic circuit generating precise switching and oscillating of multiple reagents by design, with no external control.
Figure 1: A microscale fluidic circuit generating precise switching and oscillating of multiple reagents by design, with no external control. - Video1
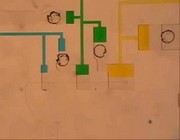 Video 1: A Fluidic Circuit that is able self-regulate the sequence and timing release of multiple reagents.
Video 1: A Fluidic Circuit that is able self-regulate the sequence and timing release of multiple reagents. - Video2
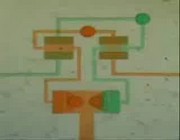 Video 2:A Fluidic Circuit capable of alternating continuously and autonomously between different states.
Video 2:A Fluidic Circuit capable of alternating continuously and autonomously between different states.
Fluid-Based Intelligent MicroCircuitsThe Problem: High-throughput biological experiments require a method to control the movement, mixing and incubation timing of microscopic amounts of reagents. While the current technology successfully miniaturized channels and chambers for these tasks, the control lines for these microchips are still very large external machines, often expensive and difficult to program and operate.
To address these problems, we have developed a new fluidic circuit technology capable to perform biological experiments on a microscale without the need for complex external machinery. Quoting the Science Daily news article on our research:
"A microfluidic device, or lab-on-a-chip, integrates multiple laboratory functions onto one chip just centimeters in size. The devices allow researchers to experiment on tiny sample sizes, and also to simultaneously perform multiple experiments on the same material...
Just as electronic circuits intelligently route the flow of electricity on computer chips without external controls, these microfluidic circuits regulate the flow of fluid through their devices without instructions from outside systems..."- Fig1
- 2010BioInfo
 Deciphering Synthetic DNA code
Deciphering Synthetic DNA code- Fig1
 Figure 1:A small section of the DNA containing encrypted information, embedded by the Craig Venter Institute into the first self-replicating synthetic bacterial cell.
Figure 1:A small section of the DNA containing encrypted information, embedded by the Craig Venter Institute into the first self-replicating synthetic bacterial cell.
Project Year:2010
Location: Univ. of Michigan, USA.
Collaborators: None.Breaking Craig Venter's Embedded Code in First Synthetic Cell.The Problem: This was not a research project in the traditional sense, but still something interesting and fun. A friend of mine pointed me to this article that describes the first synthetic living cell. That project was interesting both technologically and in terms of philosophical questions, since it shows that we can create our own design of life essentially just out of information. In the article there was a mention of an embedded encrypted code in the DNA, and a challenge to people to try to decode it.
So I decided to give it a try, got the appropriate DNA sequences (about 5000 base pairs), and a few hours later I had decyphered their new code embedded in the DNA, and found a website and email address for anybody to contact who could read messages in their DNA. After that, I got a nice email back from some people at the Venter Institute congratulating me for breaking their code and informing me that I was the 17th person in the world to have solved it so far, which is pretty good considering I didn't find out until 2 days after their original announcement so I had to start two days behind others.- Fig1
- 2005BioInfo
 Bio-Informatics / Genomics
Bio-Informatics / Genomics- Fig1
 Figure 1: New technologies enable us to reconstruct the complex interactions between genes in action rather than just learning their sequence.
Figure 1: New technologies enable us to reconstruct the complex interactions between genes in action rather than just learning their sequence. - Fig2
 Figure 2: A new technology called CAGE lets us look at more gene transcripts than ever before.
Figure 2: A new technology called CAGE lets us look at more gene transcripts than ever before. - Fig3
 Figure 3: Gene regulation starts with DNA transcription at specific sites but also continues with alternative splicing and RNA interference.
Figure 3: Gene regulation starts with DNA transcription at specific sites but also continues with alternative splicing and RNA interference.
Genomic Activity Data Analysis.The Problem: Knowing the full sequence of the Human Genome is important, but even more useful is to be able to see what parts of it are transcribed and work together at any secific time (looking at RNA instead of just DNA). To do that we relied on technology that needed us to specify both the start and end of gene transcripts. However many of our genes actually generate different splicing or alternative ending transcripts of the same gene in different situations, and would therefore go undetected.
To address these problems, the RIKEN Genomic Sciences Center in Tokyo, Japan, developed a new technique called CAGE that allowed us to look at all transcripts based just on their starting sequence, and through the work of a large consortium of international scientists, this technology was used to generate a more comlete picture of the mammalian Genome in action (the" Transcriptome"). My research revolved around the bioinformatics analysis of this new type of data.- Fig1
- 2003AI+Bio+GenEng
 Computers Re-Program Live Cells
Computers Re-Program Live Cells- Fig1
 Figure 1: Elements of Genetic Eng. by AI: cell simulation, mapping of biological parts (bacterial flagella etc) and translation into precise recombinant DNA steps.
Figure 1: Elements of Genetic Eng. by AI: cell simulation, mapping of biological parts (bacterial flagella etc) and translation into precise recombinant DNA steps. - Video1
 Video 1: Simulation of the best evolved bacterial circuit (goal: more time on red areas by sensing gradients only).
Video 1: Simulation of the best evolved bacterial circuit (goal: more time on red areas by sensing gradients only). - Video2
 Video 2: In contrast to my research, evolved A-Life creatures like this are not implementable in real biological systems, but they remain impressive demonstrations.
Video 2: In contrast to my research, evolved A-Life creatures like this are not implementable in real biological systems, but they remain impressive demonstrations.
Project Year: 1999-2003 (PhD).
Location: Univ. of Michigan, USA.
Collaborators:John H. Holland (PhD Advisor).Computer Evolution of Gene Circuits to Re-Program Live Cells.The Problem: Living cells have many useful abilities that we cannot yet reproduce in electronic or robotic systems, such as the ability to self-repair, power themselves from their environment, replicate, and generate chemicals, light, or motion at microscopic scales. They control these abilities through sets of interconnected genetic instructions called "gene circuits". Through genetic engineering we can already modify the DNA in cells and alter these gene circuits, but their complexity has limited us to small changes designed by hand rather than entire new circuits and behaviors. Computer scientists had success with much more complex systems in fields like Artificial Life, but lacked biological realism or a way to directly implement them in living systems.
To address these problems, I developed a mapping between real DNA sequences and data structures that enabled computers to simulate and evolve complex new gene circuits, and then produce a precise set of recombinant DNA procedures to embed the new gene circuits in both bacterial and human cells. My implementation takes advantage of the ability of qualitative simulations to predict overall behavior without requiring extensive and often difficult-to-measure parameters required by more quantitative simulations, and still produce robust and rapid prototyping of gene circuits from available components.- Fig1
- 2003AI+BioInfo
 Molecular Genetics Improve Computers
Molecular Genetics Improve Computers- Fig1
 Figure 1: Through computational experiments I show that search performance in computer evolution can be improved by more accurate molecular encoding.
Figure 1: Through computational experiments I show that search performance in computer evolution can be improved by more accurate molecular encoding.
Project Year: 1999-2003 (PhD).
Location: Univ. of Michigan, USA.
Collaborators:John H. Holland (PhD Advisor).Using Ideas from Molecular Genetics to Improve Artificial Intelligence.The Problem: Biological systems evolve to create clever designs for solving problems, using mechanisms that do not explicitly involve knowledge or intelligence, and yet are often superior to the best human designs. Evolution reuses and recombines previously discovered structures, building blocks, to generate more complex designs, reducing exponential problems to simpler hierarchical problems. AI search techniques like Genetic Algorithms attempt to emulate this mechanism and harness it for automated programming and problem solving and have been successfully applied in a vast number of areas as a powerful black-box design and optimization tool.
But current implementations still require significant human input in the initial problem formulation, whereas real evolution uses the same structures and mechanisms (DNA) to solve problems as different as flying or optimizing metabolic reactions.
To address these problems, I examined more carefully the molecular genetics encoding of living cells and identified and tested key mechanisms and features responsible for the success of evolution from a computational perspective. I then used them to further AI theory on evolutionary computation and build a single all-purpose implementation that does not require human intervention to setup each problem and instead uses the same sub-symbolic building blocks (gates and circuit patterns like loops, counters) for all possible problems.- Fig1
- 2001Aging+Bio+GenEng
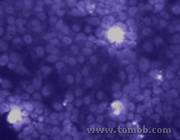 Genetic Engineering to Slow Cells' Aging
Genetic Engineering to Slow Cells' Aging - Fig1
 Figure 1: The brighter cells are secreting a fusion of immortalizing and viral proteins, which spreads into the nucleus of the surrounding normal cells.
Figure 1: The brighter cells are secreting a fusion of immortalizing and viral proteins, which spreads into the nucleus of the surrounding normal cells.
Project Year: 2001.
Location: Univ. of Michigan, USA.
Collaborators:Howard Hughes Medical Institute Lab.Interrupting Cellular Aging with Translocating Nuclear Proteins.The Problem:Normal human cells age and eventually stop growing and reproducing, reaching a state known as senescence. There are however special situations where this aging process is undone or bypassed, for example in certain cancers and in the process of making offsprings. Being able to stop cells from aging could be very useful even when considering simple applications like tissue engineering and stem cell therapies, where we might need to grow large amounts of cells that are however limited in lifespan. But while we have long known how to make cells immortal, this is done by permanently altering genes that also turn those cells into cancer cells, and therefore can no longer be used in humans.
To address these problems, I explored ways to create a similar immortalization effect in a temporary and reversible way without direct genetic modifications, by fusing these genes to viral genes capable of transporting nuclear proteins into the nucleus of new cells. These proteins would activate the same cellular immortalization pathways in the new cells, but would degraded and disappear after a few days, returning the cell to its normal state.- Fig1
- 2008Bio+Micro
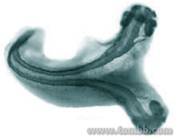 Altering ZebraFish in MicroChips
Altering ZebraFish in MicroChips- Fig1
 Figure 1: An example of double-headed fish caused by altering embryonal development.
Figure 1: An example of double-headed fish caused by altering embryonal development. - Fig2
 Figure 2: One of the zebrafish embryos grown within our microdevice, showing healthy development.
Figure 2: One of the zebrafish embryos grown within our microdevice, showing healthy development.
Project Year: 2008-2009.
Location: Univ.of Michigan, USA.
Collaborators:Micro/Mol. Lab and Barald (Neuronal Dev.) Lab.Growing Altered ZebraFish in microchipsThe Problem: Zebrafish is a small fish, just a few centimeters in length, that has been used extensively to study vertebrate development. It matures from egg to larva in just 3 days and its morphogenesis can be altered just by adding specific growth factors directly to the surrounding water, which however produces a uniform distribution in the embryo. Non-uniform distribution would be desirable as it has been used in other organisms to produce altered body development like flies with two sets of wings or two heads. The tools to direct specific growth factors or gene regulators just to specific target systems in an embryo in vertebrates rather than insects however had not been developed yet.
To address these problems, and as part of a student design project, we mentored students to design a microfluidic system to deliver cytokines to only selected parts of a zebrafish embryo, and demonstrated not only that such as system can be produced and used to grow viable adult vertebrates but also that this type of project can be used to teach students about both biology and engineering principles.- Fig1
- 2009Micro+Neuro
 C-Nanotubes in Neural Interfaces
C-Nanotubes in Neural Interfaces- Fig1
 Figure 1: Neuron cells I cultured (green), growing on the silicon substrate of the UM neural probe.
Figure 1: Neuron cells I cultured (green), growing on the silicon substrate of the UM neural probe. - Fig2
 Figure 2: I verified that Carbon nanotubes improve the interface between cells and circuitry of the neural probe.
Figure 2: I verified that Carbon nanotubes improve the interface between cells and circuitry of the neural probe. - Fig3
 Figure 3: Neurons I cultured, shown on a transparent substrate.
Figure 3: Neurons I cultured, shown on a transparent substrate.
Project Year: 2008-2009.
Location: Univ.of Michigan, USA
Collaborators:SSEL lab and Integrated Micro-Systems Lab.Interfacing Electronic Circuits with Neuron Cells using Carbon Nanotubes.The Problem: Advances in neuroscience depend on our ability to measure complex neural patterns through microscopic implantable electronic neural probes. The University of Michigan Wireless Integrated Microsystems Center has been at the forefront in this type of research. However these devices are not very compatible with brain tissue and neural cells tend to suffer damage at the interface with the probe even with adhesion protein coating.
To address these problems, I tested the effect of using carbon nanotubes (grown at one of the University's laboratories) as a coating and interface material between neuron cells and the neural probe. The results show that carbon nanotubes produced a better interface between cells and the UM's neural probe.- Fig1
- 1997AI+Robotics
 Evolved Robotics
Evolved Robotics- Fig1
 Figure 1:How to best control various body shapes in different robots is not trivial and is often best left to computer evolution.
Figure 1:How to best control various body shapes in different robots is not trivial and is often best left to computer evolution. - Video1
 Video 1: Example: evolved behaviors from another researcher, a robotic spider with evolved self-balancing (it is not glued, it just 'surfs')
Video 1: Example: evolved behaviors from another researcher, a robotic spider with evolved self-balancing (it is not glued, it just 'surfs')
Evolving Smarter Robotic Controllers.The Problem: Programming even just the motor controls of actual robots can be quite complex, as it involves mapping various sensors and coordinating appropriate responses as well-timed and precisely coordinated movements in various robotic motor units. There are different aproaches to this, including complex muathematical analysis in 3D space. A simpler and often effective approach however is to let a computer evolve the ideal control behavior within a virtual simulation until an effective solution is found. Think of newborn baby trying to learn how to move around and walk, trying many different ways over time and learning which movements work better and how much force to apply to balance, except that the baby is a robot and instead of waiting years it tries millions of simulations within just hours. This approach however can run into problems depending on how the simulation and evolution is done, for example creating solutions that seem to work within the simplified testing environment but then break down in more realistic ones.
To address these problems, we evolved a robotic controller to follow walls within a room and showed it to be much more robust, compact and effective than those evolved by previous groups, thanks to specific details in the way we handled the simulation, evolution criteria and evolution components for evolved robotic controllers.- Fig1
- 1997AI+Image
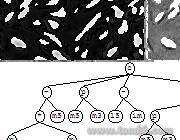 Automated AI Image Analysis
Automated AI Image Analysis- Fig1
 Figure 1: This technology was successfully applied to the automated analysis of large amounts of satellite data for geoscience research.
Figure 1: This technology was successfully applied to the automated analysis of large amounts of satellite data for geoscience research. - F2
 Figure 2: My software implementation was tested on geoscience satellite images and its results were validated against ground data and human analysis.
Figure 2: My software implementation was tested on geoscience satellite images and its results were validated against ground data and human analysis. - F3
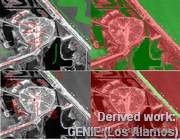 Figure 3: This project later inspired similar technologies, like the GENIE project from Los Alamos National Laboratory, here identifying streets or military targets by example.
Figure 3: This project later inspired similar technologies, like the GENIE project from Los Alamos National Laboratory, here identifying streets or military targets by example. - F4
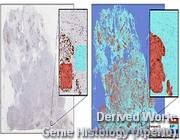 Figure 4: Technology inspired by our research is even used for commercial automated analysis of histological sections of tumors by example and then automatically.
Figure 4: Technology inspired by our research is even used for commercial automated analysis of histological sections of tumors by example and then automatically. - Video1
 Video 1: Shown an example of what to call 'edges' in an image, the AI evolves algorithms to recognize them automatically.
Video 1: Shown an example of what to call 'edges' in an image, the AI evolves algorithms to recognize them automatically.
Artificial Intelligence that Learns by Example to Analyze Image Data.The Problem: Imaging systems on satellites, telescopes and microscopes collect massive amounts of information that requires expert human eyes to interpret correctly, yet we collect far more information than experts could ever analyze on their own. But while automating this interpretation process would be desirable, experts are generally required because there is no simple rule that can replace expert interpretation. If you asked some art critics how they can measure the quality of a painting for example you are not likely to be able to write an algorithm based on their responses.
To address these problems, we designed a system that used artificial intelligence to write automated image classification algorithms not based on rules but based on examples. Take an image, ask someone to identify correctly certain features on it, and our system would evolve algorithms until they were able to take the same image and identify it in the same way as the user. Those evolved algorithms could then be used on thousands of other images to identify the same features in them.- Fig1
- 1996AI+Music
 Computers Composing Music
Computers Composing Music- Fig1
 Figure 1:In the background is one of the best performing musical compositions created through computer evolution without human interference.
Figure 1:In the background is one of the best performing musical compositions created through computer evolution without human interference.
Computers Composing Classical Music.The Problem: Music has often been considered strictly the domain of humans, and computers are often not considered 'creative' enough to compose music. Yet computers are clearly capable of finding creative solutions in areas like engineering design and problem-solving in general. In addition we can often see music that is generated by what looks like straightforward formulas, so it should be possible to obtain at least a similar level of creativity and tolerable melody from computers and artificial intelligence.
To address these problems, we used computer evolution and combined it with rules to score the partially random but continuously improving evolved output (rather than having human input, which would also work but would be much much more time-consuming). For that purpose we used the rules of 16th century classical counterpoint so that we would obtain solutions that scored better as they satisfied more and more of the rules of melodic arrangement and counterpoint.- Fig1
- 2009Micro+Laser
 Photonic Bio-Molecule Detection
Photonic Bio-Molecule Detection- Fig1
 Figure 1: Molecules binding to a substrate can cause slight but measurable deflections to laser beams.
Figure 1: Molecules binding to a substrate can cause slight but measurable deflections to laser beams. - Fig2
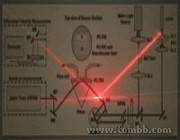 Figure 2: Our system combines microfluidics, photonic-crystals and lasers to accurately measure biomolecules.
Figure 2: Our system combines microfluidics, photonic-crystals and lasers to accurately measure biomolecules.
Project Year: 2008-2009.
Location: Univ.of Michigan, USA.
Collaborators:Ultrafast Optical Sciences Lab.Photonic Biomolecule DetectionThe Problem: Detecting the presence of small amounts of complex molecules is both an important and difficult task. Often this is done by attaching other molecules to them that are either fluorescent or magnetic particles, but even more useful are methods that are label-free, and can detect molecules without modifying them by permanently binfding something to them and interfering with later tests. A very successful technique has relied on changes in gold-film surface plasmon resonance to achieve this, but these systems are very expensive and their sensitivity could be further improved. In addition it is also useful to be able to measure not just binding but also the dissociation rates of molecules in their natural state.
To address these problems, we have developed a low-cost and highly-sensitive biomolecular detection mechanism that uses lasers and a photonic crystal in a microfluidic device, which enables for much more sensitive measurements of molecules based on the refractive index changes around the detection field.- Fig1
- 2005Quantum+Crypto.
 Unforgeable IDs with Quantum Physics
Unforgeable IDs with Quantum Physics - Fig1
 Figure 1: Documents, fingerprints, money can be forged, but Quantum Mechanics can be used to overcome this problem.
Figure 1: Documents, fingerprints, money can be forged, but Quantum Mechanics can be used to overcome this problem.
Making Unforgeable Cryptography Certificates with Quantum Physics.The Problem:With enough resources, all our current ID documents and even biometrics can be forged. And making documents that are increasingly difficult to copy or forge requires increasing effort (holograms etc). But there is a physical limit to what can be copied and how accurately when we enter the realm of quantum mechanics, and these limitations could be used there are specific limitations to what cen be done in the physical world Text
To address these problems, I considered the ultimate limits to what can be physically done to copy something, which is given by Quantum Mechanical laws. This concept has been already used successfully to build working systems that perform Quantum Cryptography, which is the transmission of information across a communication channel in a manner that cannot be intercepted. My research instead considered using quantum uncertainty and chaotic formations to make complex materials with unique and unforgeable properties that could be used as certificates.- Fig1
- 2008Bio+Micro
 Cells Mechano-Biology
Cells Mechano-Biology- Fig1
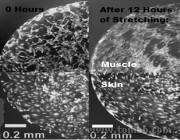
 Figure 1: The effect of periodic cycle microstretching on cell organization and alignment, in muscle and skin cells.
Figure 1: The effect of periodic cycle microstretching on cell organization and alignment, in muscle and skin cells. - Fig2
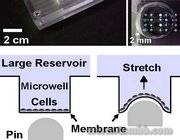 Figure 2: The mechanism of our programmable surface cell microstretcher.
Figure 2: The mechanism of our programmable surface cell microstretcher. - Fig3
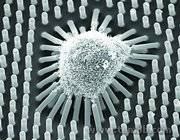 Figure 3: An example of why mechanical signals are important (from another lab1): Stem cells grappling softer microposts will change into different tissue types.
Figure 3: An example of why mechanical signals are important (from another lab1): Stem cells grappling softer microposts will change into different tissue types.
Cells Aligning to Mechanical Signals.The Problem: Cells in the body are not static and they are not isolated. Instead, they sit anchored to other tissues of various elasticity and rigidity in complex 3D structures. They expect to be periodically stretched or contracted as part of their nature. But conventional cell biology just isolates them inside static and non-physiological flasks.
One example of why this is a big problem is the recent discovery that stem cells, useful for their potential to regrow and replace damaged tissue, will actually change into different cell types depending on the type of mechanical signals they receive from their surroundings.
To address these problems, and to recreate these mechanical signals, we built a microdevice capable of exposing cells to very complex patterns of mechanical stretching over time, and observed different amounts of cell organization and restructuring as a result.- Fig1
- 2010Bio+Micro
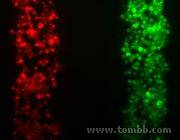 Tissue Engineering & Cell Migration
Tissue Engineering & Cell Migration- Fig1
 Figure 1:We positioned different 'columns' of cells, with one cell type generating or absorbing gradients of chemo-attractants to guide the other.
Figure 1:We positioned different 'columns' of cells, with one cell type generating or absorbing gradients of chemo-attractants to guide the other. - Fig2
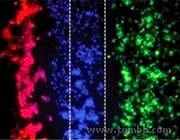 Figure 2:We can measure the movement of certain cells (in blue), like stem cells or cancer cells, toward specific tissues (red vs green).
Figure 2:We can measure the movement of certain cells (in blue), like stem cells or cancer cells, toward specific tissues (red vs green). - Fig3
 Figure 3:An example of more complex patterning (from a different research project in our lab) using the same method used in our chemotaxis experiment.
Figure 3:An example of more complex patterning (from a different research project in our lab) using the same method used in our chemotaxis experiment.
Controlling Cell Migration with MicroGradients and Multiple Cell Types.The Problem: Cells in our body do not always stay in one place. During infection, cancer, healing and even during fetal development or growth, different cells send signals to each other to migrate to different locations. In the body, the signals spread slowly through tissues and form a gradient (higher concentration near the source cell) that tells cells in which direction to go. These signals are important (modifying them can generate creatures with abnormal development of body structures, block or facilitate cancer or muscle growth etc), but they have been difficult to study with conventional means (dishes of cells) because these signals would just get diluted and diffuse out rather than forming gradients that cells could sense and follow.
To address these problems, we developed microscopic devices capable of patterning cells to generate gradients inside microchannels and then observe and accurately measure the effect of different factors, including more complex gradients formed by both generator and sink cells for these signals.- Fig1
- 10 Scientific Journals
- 2011 Lab.Chip
 Cited by 05Journals
Cited by 05Journals 2011: Lab on a ChipMosadegh, B., Bersano-Begey, T., Park, JY., Burns, Mark, and Takayama S. (2011). "Next-generation integrated microfluidic circuits.", Lab on a Chip, 2011, 11, 2813-2818 DOI: 10.1039/C1LC20387HAbstract:This mini-review provides a brief overview of recent devices that use networks of elastomeric valves to minimize or eliminate the need for interconnections between microfluidic chips and external instruction lines that send flow control signals. Conventional microfluidic control mechanisms convey instruction signals in a parallel manner such that the number of instruction lines must increase as the number of independently operated valves increases. The devices described here circumvent this "tyranny of microfluidic interconnects" by the serial encoding of information to enable instruction of an arbitrary number of independent valves with a set number of control lines, or by the microfluidic circuit-embedded encoding of instructions to eliminate control lines altogether. Because the parallel instruction chips are the most historical and straightforward to design, they are still the most commonly used approach today. As requirements for instruction complexity, chip-to-chip communication, and real-time on-chip feedback flow control arise, the next generation of integrated microfluidic circuits will need to incorporate these latest interconnect flow control approaches.
2011: Lab on a ChipMosadegh, B., Bersano-Begey, T., Park, JY., Burns, Mark, and Takayama S. (2011). "Next-generation integrated microfluidic circuits.", Lab on a Chip, 2011, 11, 2813-2818 DOI: 10.1039/C1LC20387HAbstract:This mini-review provides a brief overview of recent devices that use networks of elastomeric valves to minimize or eliminate the need for interconnections between microfluidic chips and external instruction lines that send flow control signals. Conventional microfluidic control mechanisms convey instruction signals in a parallel manner such that the number of instruction lines must increase as the number of independently operated valves increases. The devices described here circumvent this "tyranny of microfluidic interconnects" by the serial encoding of information to enable instruction of an arbitrary number of independent valves with a set number of control lines, or by the microfluidic circuit-embedded encoding of instructions to eliminate control lines altogether. Because the parallel instruction chips are the most historical and straightforward to design, they are still the most commonly used approach today. As requirements for instruction complexity, chip-to-chip communication, and real-time on-chip feedback flow control arise, the next generation of integrated microfluidic circuits will need to incorporate these latest interconnect flow control approaches. - 2011 AF.Mat.
 Cited by 02Journals
Cited by 02Journals 2010: Adv. Functional MaterialsTavana, H., Kaylan, K, Bersano-Begey, T., Luker, KE., Luker, GD., Takayama, S. (2011). "Rehydration of Polymeric, Aqueous, Biphasic System Facilitates High Throughput Cell Exclusion Patterning for Cell Migration Studies.", Advanced Functional Materials Volume 21, Issue 15, pages 2920-2926, August 9, 2011 DOI: 10.1002/adfm.201002559Abstract:This paper describes a cell-exclusion patterning method facilitated by a polymeric aqueous two-phase system. The immersion aqueous phase (polyethylene glycol) containing cells rehydrates a dried disk of the denser phase (dextran) on the substrate to form a dextran droplet. With the right properties of the phase-forming polymers, the rehydrating droplet remains immiscible with the immersion phase. Proper formulation of the two-phase system ensures that the interfacial tension between the rehydrating droplet and the surrounding aqueous phase prevents cells from crossing the interface so that cells only adhere to the regions of the substrate around the dextran phase droplet. Washing out the patterning two-phase reagents reveals a cell monolayer containing a well-defined circular gap that serves as the migration niche for cells of the monolayer. Migration of cells into the cell-excluded area is readily visualized and quantified over time. A 96-well plate format of this "gap healing" migration assay demonstrates the ability to detect inhibition of cell migration by known cytoskeleton targeting agents. This straightforward method, which only requires a conventional liquid handler and readily prepared polymer solutions, opens new opportunities for high throughput cell migration assays.
2010: Adv. Functional MaterialsTavana, H., Kaylan, K, Bersano-Begey, T., Luker, KE., Luker, GD., Takayama, S. (2011). "Rehydration of Polymeric, Aqueous, Biphasic System Facilitates High Throughput Cell Exclusion Patterning for Cell Migration Studies.", Advanced Functional Materials Volume 21, Issue 15, pages 2920-2926, August 9, 2011 DOI: 10.1002/adfm.201002559Abstract:This paper describes a cell-exclusion patterning method facilitated by a polymeric aqueous two-phase system. The immersion aqueous phase (polyethylene glycol) containing cells rehydrates a dried disk of the denser phase (dextran) on the substrate to form a dextran droplet. With the right properties of the phase-forming polymers, the rehydrating droplet remains immiscible with the immersion phase. Proper formulation of the two-phase system ensures that the interfacial tension between the rehydrating droplet and the surrounding aqueous phase prevents cells from crossing the interface so that cells only adhere to the regions of the substrate around the dextran phase droplet. Washing out the patterning two-phase reagents reveals a cell monolayer containing a well-defined circular gap that serves as the migration niche for cells of the monolayer. Migration of cells into the cell-excluded area is readily visualized and quantified over time. A 96-well plate format of this "gap healing" migration assay demonstrates the ability to detect inhibition of cell migration by known cytoskeleton targeting agents. This straightforward method, which only requires a conventional liquid handler and readily prepared polymer solutions, opens new opportunities for high throughput cell migration assays. - 2010 Nat.Phys.
 Cited by 33Journals
Cited by 33Journals 2010: Nature PhysicsMosadegh, B., Kuo, C.H., Tung, Y.C., Torisawa, Y., Bersano-Begey, T., Tavana, H., and Takayama S. (2010). "Integrated elastomeric components for autonomous control of microfluidic switching and oscillation", Nature Physics, 6 (2010) 433-437.Abstract:A critical need for enhancing usability and capabilities of microfluidic technologies is the development of standardized, scalable, and versatile control systems1,2. Electronically controlled valves and pumps typically used for dynamic flow regulation, although useful, can limit convenience, scalability, and robustness3-5. This shortcoming has motivated development of device-embedded non-electrical flow-control systems. Existing approaches to regulate operation timing on-chip, however, still require external signals such as timed generation of fluid flow, bubbles, liquid plugs or droplets, or an alteration of chemical compositions or temperature6-16. Here, we describe a strategy to provide device-embedded flow switching and clocking functions. Physical gaps and cavities interconnected by holes are fabricated into a three-layer elastomer structure to form networks of fluidic gates that can spontaneously generate cascading and oscillatory flow output using only a constant flow of Newtonian fluids as the device input. The resulting microfluidic substrate architecture is simple, scalable, and should be applicable to various materials. This flow-powered fluidic gating scheme brings the autonomous signal processing ability of microelectronic circuits to microfluidics where there is the added diversity in current information of having distinct chemical or particulate species and richness in current operation of having chemical reactions and physical interactions.
2010: Nature PhysicsMosadegh, B., Kuo, C.H., Tung, Y.C., Torisawa, Y., Bersano-Begey, T., Tavana, H., and Takayama S. (2010). "Integrated elastomeric components for autonomous control of microfluidic switching and oscillation", Nature Physics, 6 (2010) 433-437.Abstract:A critical need for enhancing usability and capabilities of microfluidic technologies is the development of standardized, scalable, and versatile control systems1,2. Electronically controlled valves and pumps typically used for dynamic flow regulation, although useful, can limit convenience, scalability, and robustness3-5. This shortcoming has motivated development of device-embedded non-electrical flow-control systems. Existing approaches to regulate operation timing on-chip, however, still require external signals such as timed generation of fluid flow, bubbles, liquid plugs or droplets, or an alteration of chemical compositions or temperature6-16. Here, we describe a strategy to provide device-embedded flow switching and clocking functions. Physical gaps and cavities interconnected by holes are fabricated into a three-layer elastomer structure to form networks of fluidic gates that can spontaneously generate cascading and oscillatory flow output using only a constant flow of Newtonian fluids as the device input. The resulting microfluidic substrate architecture is simple, scalable, and should be applicable to various materials. This flow-powered fluidic gating scheme brings the autonomous signal processing ability of microelectronic circuits to microfluidics where there is the added diversity in current information of having distinct chemical or particulate species and richness in current operation of having chemical reactions and physical interactions. - 2010 Int.Bio
 Cited by 10Journals
Cited by 10Journals 2010: Integrative BiologyTorisawa YS, Mosadegh B, Bersano-Begey T, Steele JM, Luker KE, Luker GD, Takayama S. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Journal of Integrative Biology, 2010, 2 (11), 680 - 686.Abstract:Chemokine CXCL12 promotes CXCR4-dependent chemotaxis of cancer cells to characteristic organs and tissues, leading to metastatic disease. This study was designed to investigate how cells expressing CXCR7 regulate chemotaxis of a separate population of CXCR4 cells under physiologic conditions in which cells are exposed to gradients of CXCL12. We recapitulated a cancer-stroma microenvironment by patterning CXCR4-expressing cancer cells in microchannels at spatially defined positions relative to CXCL12-producing cells and CXCR7-expressing cells. CXCR7 scavenges and degrades CXCL12, which has been proposed to facilitate CXCR4-dependent chemotaxis through a source-sink model. Using the microchannel device, we demonstrated that chemotaxis of CXCR4 cells depended critically on the presence and location of CXCR7 cells (sink) relative to chemokine secreting cells (source). Furthermore, inhibiting CXCR4 on migrating cells or CXCR7 on sink cells blocked CXCR4-dependent chemotaxis toward CXCL12, showing that the device can identify new therapeutic agents that block migration by targeting chemoattractant scavenging receptors. Our system enables efficient chemotaxis under much shallower yet more physiological chemoattractant gradients by generating an in vitro microenvironment where combinations of cellular products may be secreted along with formation of a chemoattractant gradient. In addition to elucidating mechanisms of CXCL-12 mediated chemotaxis, this simple and robust method can be broadly useful for engineering multiple microenvironments to investigate intercellular communication.
2010: Integrative BiologyTorisawa YS, Mosadegh B, Bersano-Begey T, Steele JM, Luker KE, Luker GD, Takayama S. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Journal of Integrative Biology, 2010, 2 (11), 680 - 686.Abstract:Chemokine CXCL12 promotes CXCR4-dependent chemotaxis of cancer cells to characteristic organs and tissues, leading to metastatic disease. This study was designed to investigate how cells expressing CXCR7 regulate chemotaxis of a separate population of CXCR4 cells under physiologic conditions in which cells are exposed to gradients of CXCL12. We recapitulated a cancer-stroma microenvironment by patterning CXCR4-expressing cancer cells in microchannels at spatially defined positions relative to CXCL12-producing cells and CXCR7-expressing cells. CXCR7 scavenges and degrades CXCL12, which has been proposed to facilitate CXCR4-dependent chemotaxis through a source-sink model. Using the microchannel device, we demonstrated that chemotaxis of CXCR4 cells depended critically on the presence and location of CXCR7 cells (sink) relative to chemokine secreting cells (source). Furthermore, inhibiting CXCR4 on migrating cells or CXCR7 on sink cells blocked CXCR4-dependent chemotaxis toward CXCL12, showing that the device can identify new therapeutic agents that block migration by targeting chemoattractant scavenging receptors. Our system enables efficient chemotaxis under much shallower yet more physiological chemoattractant gradients by generating an in vitro microenvironment where combinations of cellular products may be secreted along with formation of a chemoattractant gradient. In addition to elucidating mechanisms of CXCL-12 mediated chemotaxis, this simple and robust method can be broadly useful for engineering multiple microenvironments to investigate intercellular communication. - 2010 Lab.Chip
 Cited by 05Journals
Cited by 05Journals 2010: Lab on a ChipMosadegh B, Agarwal M, Tavana H, Bersano-Begey T, Torisawa YS, Morell M, Wyatt MJ, O'Shea KS, Barald KF, Takayama S. Uniform cell seeding and generation of overlapping gradient profiles in a multiplexed microchamber device with normally-closed valves. Lab on a Chip Journal. 2010 Sep 9.Abstract:Generation of stable soluble-factor gradients in microfluidic devices enables studies of various cellular events such as chemotaxis and differentiation. However, many gradient devices directly expose cells to constant fluid flow and that can induce undesired responses from cells due to shear stress and/or wash out of cell-secreted molecules. Although there have been devices with flow-free gradients, they typically generate only a single condition and/or have a decaying gradient profile that does not accommodate long-term experiments. Here we describe a microdevice that generates several chemical gradient conditions on a single platform in flow-free microchambers which facilitates steady-state gradient profiles. The device contains embedded normally-closed valves that enable fast and uniform seeding of cells to all microchambers simultaneously. A network of microchannels distributes desired solutions from easy-access open reservoirs to a single output port, enabling a simple setup for inducing flow in the device. Embedded porous filters, sandwiched between the microchannel networks and cell microchambers, enable diffusion of biomolecules but inhibit any bulk flow over the cells.
2010: Lab on a ChipMosadegh B, Agarwal M, Tavana H, Bersano-Begey T, Torisawa YS, Morell M, Wyatt MJ, O'Shea KS, Barald KF, Takayama S. Uniform cell seeding and generation of overlapping gradient profiles in a multiplexed microchamber device with normally-closed valves. Lab on a Chip Journal. 2010 Sep 9.Abstract:Generation of stable soluble-factor gradients in microfluidic devices enables studies of various cellular events such as chemotaxis and differentiation. However, many gradient devices directly expose cells to constant fluid flow and that can induce undesired responses from cells due to shear stress and/or wash out of cell-secreted molecules. Although there have been devices with flow-free gradients, they typically generate only a single condition and/or have a decaying gradient profile that does not accommodate long-term experiments. Here we describe a microdevice that generates several chemical gradient conditions on a single platform in flow-free microchambers which facilitates steady-state gradient profiles. The device contains embedded normally-closed valves that enable fast and uniform seeding of cells to all microchambers simultaneously. A network of microchannels distributes desired solutions from easy-access open reservoirs to a single output port, enabling a simple setup for inducing flow in the device. Embedded porous filters, sandwiched between the microchannel networks and cell microchambers, enable diffusion of biomolecules but inhibit any bulk flow over the cells. - 2009 Zebrafish
 Cited by 06Journals
Cited by 06Journals 2009: ZebrafishShen, YC; Li, D; Al-Shoaibi, A; Bersano-Begey, T; Chen, H; Ali, S; Flak, S; Perrin, C; Winslow, M; Shah, H; Ramamurthy, P; Schmedlen, RH; Takayama, S; Barald, K; A Student Team in a University of Michigan Biomedical Engineering Design Course Constructs a Microfluidic Bioreactor for Studies of Zebrafish Development Zebrafish. June 2009, 6(2): 201-213.Abstract:The zebrafish is a valuable model for teaching developmental, molecular, and cell biology; aquatic sciences; comparative anatomy; physiology; and genetics. Here we demonstrate that zebrafish provide an excellent model system to teach engineering principles. A seven-member undergraduate team in a biomedical engineering class designed, built, and tested a zebrafish microfluidic bioreactor applying microfluidics, an emerging engineering technology, to study zebrafish development. During the semester, students learned engineering and biology experimental design, chip microfabrication, mathematical modeling, zebrafish husbandry, principles of developmental biology, fluid dynamics, microscopy, and basic molecular biology theory and techniques. The team worked to maximize each person's contribution and presented weekly written and oral reports. Two postdoctoral fellows, a graduate student, and three faculty instructors coordinated and directed the team in an optimal blending of engineering, molecular, and developmental biology skill sets. The students presented two posters, including one at the Zebrafish meetings in Madison, Wisconsin (June 2008).
2009: ZebrafishShen, YC; Li, D; Al-Shoaibi, A; Bersano-Begey, T; Chen, H; Ali, S; Flak, S; Perrin, C; Winslow, M; Shah, H; Ramamurthy, P; Schmedlen, RH; Takayama, S; Barald, K; A Student Team in a University of Michigan Biomedical Engineering Design Course Constructs a Microfluidic Bioreactor for Studies of Zebrafish Development Zebrafish. June 2009, 6(2): 201-213.Abstract:The zebrafish is a valuable model for teaching developmental, molecular, and cell biology; aquatic sciences; comparative anatomy; physiology; and genetics. Here we demonstrate that zebrafish provide an excellent model system to teach engineering principles. A seven-member undergraduate team in a biomedical engineering class designed, built, and tested a zebrafish microfluidic bioreactor applying microfluidics, an emerging engineering technology, to study zebrafish development. During the semester, students learned engineering and biology experimental design, chip microfabrication, mathematical modeling, zebrafish husbandry, principles of developmental biology, fluid dynamics, microscopy, and basic molecular biology theory and techniques. The team worked to maximize each person's contribution and presented weekly written and oral reports. Two postdoctoral fellows, a graduate student, and three faculty instructors coordinated and directed the team in an optimal blending of engineering, molecular, and developmental biology skill sets. The students presented two posters, including one at the Zebrafish meetings in Madison, Wisconsin (June 2008). - 2008 BioMat.
 Cited by 26Journals
Cited by 26Journals 2008: BiomaterialsKamotani, Y., Bersano-Begey, T., Kato, N., Tung, Y.C., Huh, D., Song, J.W., and Takayama, S. (2008). Individually programmable cell stretching microwell arrays actuated by a Braille display. Biomaterials 29, 2646-2655.Abstract:Cell culture systems are often static and are therefore nonphysiological. In vivo, many cells are exposed to dynamic surroundings that stimulate cellular responses in a process known as mechanotransduction. To recreate this environment, stretchable cell culture substrate systems have been developed, however, these systems are limited by being macroscopic and low throughput. We have developed a device consisting of 24 miniature cell stretching chambers with flexible bottom membranes that are deformed using the computer-controlled, piezoelectrically actuated pins of a Braille display. We have also developed efficient image capture and analysis protocols to quantify morphological responses of the cells to applied strain. Human dermal microvascular endothelial cells (HDMECs) were found to show increasing degrees of alignment and elongation perpendicular to the radial strain in response to cyclic stretch at increasing frequencies of 0.2, 1, and 5 Hz, after 2, 4, and 12h. Mouse myogenic C2C12 cells were also found to align in response to the stretch, while A549 human lung adenocarcinoma epithelial cells did not respond to stretch.
2008: BiomaterialsKamotani, Y., Bersano-Begey, T., Kato, N., Tung, Y.C., Huh, D., Song, J.W., and Takayama, S. (2008). Individually programmable cell stretching microwell arrays actuated by a Braille display. Biomaterials 29, 2646-2655.Abstract:Cell culture systems are often static and are therefore nonphysiological. In vivo, many cells are exposed to dynamic surroundings that stimulate cellular responses in a process known as mechanotransduction. To recreate this environment, stretchable cell culture substrate systems have been developed, however, these systems are limited by being macroscopic and low throughput. We have developed a device consisting of 24 miniature cell stretching chambers with flexible bottom membranes that are deformed using the computer-controlled, piezoelectrically actuated pins of a Braille display. We have also developed efficient image capture and analysis protocols to quantify morphological responses of the cells to applied strain. Human dermal microvascular endothelial cells (HDMECs) were found to show increasing degrees of alignment and elongation perpendicular to the radial strain in response to cyclic stretch at increasing frequencies of 0.2, 1, and 5 Hz, after 2, 4, and 12h. Mouse myogenic C2C12 cells were also found to align in response to the stretch, while A549 human lung adenocarcinoma epithelial cells did not respond to stretch. - 2007 BioMed.
 Cited by 69Journals
Cited by 69Journals 2007: Biomedical MicrodevicesMehta, G., Mehta, K., Sud, D., Song, J.W., Bersano-Begey, T., Futai, N., Heo, Y.S., Mycek, M.A., Linderman, J.J., and Takayama, S. (2007). Quantitative measurement and control of oxygen levels in microfluidic poly(dimethylsiloxane) bioreactors during cell culture. Biomedical microdevices 9, 123-134.Abstract:Microfluidic bioreactors fabricated from highly gas-permeable poly(dimethylsiloxane) (PDMS) materials have been observed, somewhat unexpectedly, to give rise to heterogeneous long term responses along the length of a perfused mammalian cell culture channel, reminiscent of physiologic tissue zonation that arises at least in part due to oxygen gradients. To develop a more quantitative understanding and enable better control of the physical-chemical mechanisms underlying cell biological events in such PDMS reactors, dissolved oxygen concentrations in the channel system were quantified in real time using fluorescence intensity and lifetime imaging of an oxygen sensitive dye, ruthenium tris(2,2'-dipyridyl) dichloride hexahydrate (RTDP). The data indicate that despite oxygen diffusion through PDMS, uptake of oxygen by cells inside the perfused PDMS microchannels induces an axial oxygen concentration gradient, with lower levels recorded in downstream regions. The oxygen concentration gradient generated by a balance of cellular uptake, convective transport by media flow, and permeation through PDMS in our devices ranged from 0.0003 (mg/l)/mm to 0.7 (mg/l)/mm. The existence of such steep gradients induced by cellular uptake can have important biological consequences. Results are consistent with our mathematical model and give insight into the conditions under which flux of oxygen through PDMS into the microchannels will or will not contribute significantly to oxygen delivery to cells and also provide a design tool to manipulate and control oxygen for cell culture and device engineering. The combination of computerized microfluidics, in situ oxygen sensing, and mathematical models opens new windows for microphysiologic studies utilizing oxygen gradients and low oxygen tensions.
2007: Biomedical MicrodevicesMehta, G., Mehta, K., Sud, D., Song, J.W., Bersano-Begey, T., Futai, N., Heo, Y.S., Mycek, M.A., Linderman, J.J., and Takayama, S. (2007). Quantitative measurement and control of oxygen levels in microfluidic poly(dimethylsiloxane) bioreactors during cell culture. Biomedical microdevices 9, 123-134.Abstract:Microfluidic bioreactors fabricated from highly gas-permeable poly(dimethylsiloxane) (PDMS) materials have been observed, somewhat unexpectedly, to give rise to heterogeneous long term responses along the length of a perfused mammalian cell culture channel, reminiscent of physiologic tissue zonation that arises at least in part due to oxygen gradients. To develop a more quantitative understanding and enable better control of the physical-chemical mechanisms underlying cell biological events in such PDMS reactors, dissolved oxygen concentrations in the channel system were quantified in real time using fluorescence intensity and lifetime imaging of an oxygen sensitive dye, ruthenium tris(2,2'-dipyridyl) dichloride hexahydrate (RTDP). The data indicate that despite oxygen diffusion through PDMS, uptake of oxygen by cells inside the perfused PDMS microchannels induces an axial oxygen concentration gradient, with lower levels recorded in downstream regions. The oxygen concentration gradient generated by a balance of cellular uptake, convective transport by media flow, and permeation through PDMS in our devices ranged from 0.0003 (mg/l)/mm to 0.7 (mg/l)/mm. The existence of such steep gradients induced by cellular uptake can have important biological consequences. Results are consistent with our mathematical model and give insight into the conditions under which flux of oxygen through PDMS into the microchannels will or will not contribute significantly to oxygen delivery to cells and also provide a design tool to manipulate and control oxygen for cell culture and device engineering. The combination of computerized microfluidics, in situ oxygen sensing, and mathematical models opens new windows for microphysiologic studies utilizing oxygen gradients and low oxygen tensions. - 2005 Science
 CitedBy1171Others
CitedBy1171Others 2005: ScienceCarninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M.C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., Kodzius, R., Shimokawa, K., Bajic, VB.,... Bersano , T., et al. (2005). The transcriptional landscape of the mammalian genome. Science, New York, NY 309, 1559-1563.Abstract:This study describes comprehensive polling of transcription start and termination sites and analysis of previously unidentified full-length complementary DNAs derived from the mouse genome. We identify the 5' and 3' boundaries of 181,047 transcripts with extensive variation in transcripts arising from alternative promoter usage, splicing, and polyadenylation. There are 16,247 new mouse protein-coding transcripts, including 5154 encoding previously unidentified proteins. Genomic mapping of the transcriptome reveals transcriptional forests, with overlapping transcription on both strands, separated by deserts in which few transcripts are observed. The data provide a comprehensive platform for the comparative analysis of mammalian transcriptional regulation in differentiation and development.
2005: ScienceCarninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M.C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., Kodzius, R., Shimokawa, K., Bajic, VB.,... Bersano , T., et al. (2005). The transcriptional landscape of the mammalian genome. Science, New York, NY 309, 1559-1563.Abstract:This study describes comprehensive polling of transcription start and termination sites and analysis of previously unidentified full-length complementary DNAs derived from the mouse genome. We identify the 5' and 3' boundaries of 181,047 transcripts with extensive variation in transcripts arising from alternative promoter usage, splicing, and polyadenylation. There are 16,247 new mouse protein-coding transcripts, including 5154 encoding previously unidentified proteins. Genomic mapping of the transcriptome reveals transcriptional forests, with overlapping transcription on both strands, separated by deserts in which few transcripts are observed. The data provide a comprehensive platform for the comparative analysis of mammalian transcriptional regulation in differentiation and development. - 2003 MedChem
 Cited by 47Journals
Cited by 47Journals 2003: Curr. Topics in Medicinal ChemistryShim, J., Bersano-Begey, T.F., Zhu, X., Tkaczyk, A.H., Linderman, J.J., and Takayama, S. (2003). Micro- and nanotechnologies for studying cellular function. Current topics in medicinal chemistry 3, 687-703.Abstract:The study of complex biological systems requires methods to perturb the system in complex yet controlled ways to elucidate mechanisms and dynamic interactions, and to recreate in vivo conditions in flexible in vitro set-ups. This paper reviews recent advances in the use of micro- and nanotechnologies in the study of complex biological systems and the advantages they provide in these two areas. Particularly useful for controlling the chemical and mechanical microenvironments of cells is a set of techniques called soft lithography, whereby elastomeric materials are used to transfer and generate micro- and nanoscale patterns. Examples of some of the capabilities of soft lithography include the use of elastomeric stamps to generate micropatterns of protein and the use of elastomeric channels to localize chemicals with subcellular spatial resolutions. These types of biological micro- and nanotechnologies combined with mathematical modeling will propel our understandings of cellular and subcellular physiology to new heights.
2003: Curr. Topics in Medicinal ChemistryShim, J., Bersano-Begey, T.F., Zhu, X., Tkaczyk, A.H., Linderman, J.J., and Takayama, S. (2003). Micro- and nanotechnologies for studying cellular function. Current topics in medicinal chemistry 3, 687-703.Abstract:The study of complex biological systems requires methods to perturb the system in complex yet controlled ways to elucidate mechanisms and dynamic interactions, and to recreate in vivo conditions in flexible in vitro set-ups. This paper reviews recent advances in the use of micro- and nanotechnologies in the study of complex biological systems and the advantages they provide in these two areas. Particularly useful for controlling the chemical and mechanical microenvironments of cells is a set of techniques called soft lithography, whereby elastomeric materials are used to transfer and generate micro- and nanoscale patterns. Examples of some of the capabilities of soft lithography include the use of elastomeric stamps to generate micropatterns of protein and the use of elastomeric channels to localize chemicals with subcellular spatial resolutions. These types of biological micro- and nanotechnologies combined with mathematical modeling will propel our understandings of cellular and subcellular physiology to new heights. - 4 Books and Invited Talks
- 2009 (Book)

 BOOK CHAPTER2009: Lab on a Chip TechnologiesBersano-Begey, T.F., Takayama, S., (2009). " Braille Microfluidics ". Lab on a Chip Technologies and Applications , Chapter 16. Eds. Rasooly A, Herold KE. Horizon Scientific Press, Norwich, UK.Abstract:Chapter 16 Braille Microfluidics: This chapter describes protocols for fabrication and assembly of microfluidic chips designed for Braille-display-based fluid actuation. Emphasis is placed on critical fabrication details such as channel cross-sectional shape and dimensions, and membrane thickness. Commercial sources for the Braille display hardware and some simple software operation are also described. A detailed example of a protocol for a chip used for immunoassays using the Braille display for fluid actuation is also described.
BOOK CHAPTER2009: Lab on a Chip TechnologiesBersano-Begey, T.F., Takayama, S., (2009). " Braille Microfluidics ". Lab on a Chip Technologies and Applications , Chapter 16. Eds. Rasooly A, Herold KE. Horizon Scientific Press, Norwich, UK.Abstract:Chapter 16 Braille Microfluidics: This chapter describes protocols for fabrication and assembly of microfluidic chips designed for Braille-display-based fluid actuation. Emphasis is placed on critical fabrication details such as channel cross-sectional shape and dimensions, and membrane thickness. Commercial sources for the Braille display hardware and some simple software operation are also described. A detailed example of a protocol for a chip used for immunoassays using the Braille display for fluid actuation is also described.
Braille Microfluidics is a microfluidic technology which uses arrays of small retractable pins on pre-existing Braille hardware as the actuation and control mechanism for elastomeric microfluidic chips. Braille actuation enables microfluidic pumping and valving without tubings and interconnects; the channels within the microfluidic chip are fabricated with a thin bottom membrane and are aligned over the Braille pins to interact with them. As the pins are raised or retracted they can perform all of the basic microfluidic operations such as valving, peristaltic pumping, and mixing.
Among the advantages of this setup are ease of fabrication (single layer and no interconnections), portability (the entire system can be configured to occupy just the size of a hand) and high-programmability (all pins, and hence all the microfluidic valves they actuate, can be independently addressed and controlled automatically through a computer with a USB connection). However the same setup also introduces new challenges, such as an increased evaporation rate through its thin bottom membrane and a pulsatile flow which is not always desirable. A few solutions to these problems along with applications in which the Braille system is used are briefly discussed. - 2006 (Book)

 BOOK CHAPTER2006: Encycl. of Medical Devices and InstrumentationZhu, X., Bersano-Begey, T., Kamotani, Y., Takayama, S., (2006). " Microbioreactors". Wiley Encyclopedia of Medical Devices and Instrumentation, 2nd edition. J.G. Webster (editior). John Wiley & Sons.Abstract:Recent advances in micro- and nanotechnologies coupled with the need in biology and medicine to perform cellular scale studies and productions have led to the emergence of a variety of microscale biological reaction systems. These microbioreactors are promising as low cost modules for optimizing bioproduction processes, highly parallel analytical platforms for screening pharmaceutical drugs, therapeutic implements for treating disorders and deficiencies, and examination tools for exploring basic physiological responses and biological reactions. This article summarizes the basic concepts, designs, and applications of microbioreactors with a special focus on medical, pharmaceutical, and cell biological applications. Important concepts include scaling laws and how the small size of the microbioreactors affects fluid flow, mixing, mass transport, heat transfer, and sterilization. Examples of specific microbioreactors for fermentation, drug toxicity screening, drug administration, assisted reproduction, and basic cell biological and physiological studies are also discussed. Microscale phenomena provide both a challenge and an opportunity for the development of microbioreactors.
BOOK CHAPTER2006: Encycl. of Medical Devices and InstrumentationZhu, X., Bersano-Begey, T., Kamotani, Y., Takayama, S., (2006). " Microbioreactors". Wiley Encyclopedia of Medical Devices and Instrumentation, 2nd edition. J.G. Webster (editior). John Wiley & Sons.Abstract:Recent advances in micro- and nanotechnologies coupled with the need in biology and medicine to perform cellular scale studies and productions have led to the emergence of a variety of microscale biological reaction systems. These microbioreactors are promising as low cost modules for optimizing bioproduction processes, highly parallel analytical platforms for screening pharmaceutical drugs, therapeutic implements for treating disorders and deficiencies, and examination tools for exploring basic physiological responses and biological reactions. This article summarizes the basic concepts, designs, and applications of microbioreactors with a special focus on medical, pharmaceutical, and cell biological applications. Important concepts include scaling laws and how the small size of the microbioreactors affects fluid flow, mixing, mass transport, heat transfer, and sterilization. Examples of specific microbioreactors for fermentation, drug toxicity screening, drug administration, assisted reproduction, and basic cell biological and physiological studies are also discussed. Microscale phenomena provide both a challenge and an opportunity for the development of microbioreactors. - 2004 (Book)

 BOOK CHAPTER2004: Encycl. of Nanoscience and NanoTechnologyZhu, X., Bersano-Begey, T., Takayama, S., (2004) " Nanomaterials for Cell Engineering". Encyclopedia of Nanoscience and Nanotechnology , Ed. Nalwa, HS, Vol 6, p 857-878. American Scientific Publishers ISBN 1-58883-001-2.Abstract:The study and control of cell function require materials with well-defined nanoscale features. In multicellular organisms, the extracellular matrix presents nanoscale geometrical features, adhesive cues, and mechanical stimuli to cells. Although it is the genetic nanomaterial inside the cell that determines the range of possible cell behaviors, it is these extracellular microand nanoenvironments that play a major role in determining which of the behavioral possibilities are realized.
BOOK CHAPTER2004: Encycl. of Nanoscience and NanoTechnologyZhu, X., Bersano-Begey, T., Takayama, S., (2004) " Nanomaterials for Cell Engineering". Encyclopedia of Nanoscience and Nanotechnology , Ed. Nalwa, HS, Vol 6, p 857-878. American Scientific Publishers ISBN 1-58883-001-2.Abstract:The study and control of cell function require materials with well-defined nanoscale features. In multicellular organisms, the extracellular matrix presents nanoscale geometrical features, adhesive cues, and mechanical stimuli to cells. Although it is the genetic nanomaterial inside the cell that determines the range of possible cell behaviors, it is these extracellular microand nanoenvironments that play a major role in determining which of the behavioral possibilities are realized.
Nanoenvironments have a large influence on cell behaviors such as cell orientation, migration, proliferation, and differentiation. Many new applications that involve cells -such as tissue engineering, studies of the cellular mechanism of diseases, and the development of cellbased therapies, cellular biosensors, and machine-cell hybrids- require engineered materials that can elicit desired responses from contacting cells.
This chapter starts with a brief overview of the nanoscale characteristic of the cells and how they "read" extracellular cues presented by nanomaterials. The chapter then summarizes recent efforts in creating nanomaterials for engineering cells in three sections that focus on the three different aspects of nanoscale features: geometry, chemistry, and mechanics. - 1996 (Book)
 Cited by 45Journals
Cited by 45Journals BOOK CHAPTER1996: Adv. in Genetic Programming 2Daida, J.M., Hommes, J.D., Bersano-Begey, T., Ross, S.J., Vesecky, J.F., (1996)." Algorithm Discovery Using the Genetic Programming Paradigm: Extracting Low-Contrast Curvilinear Features from SAR Images of Arctic Ice". Advances in Genetic Programming II , P. Angeline and K. Kinnear, Jr. (ed.) Cambridge: The MIT Press. Cited by 38 other publications.Abstract:This chapter discusses the application of genetic programming (GP) to image analysis problems in geoscience and remote sensing and describes how a GP can be adapted for processing large data sets (in our case, 1024 x 1024 pixel images plus texture channels). The featured problem is one that has not been adequately solved for this type of imagery. We describe the placement of GP in the overall scheme of algorithm discovery in geoscience image analysis and describe how GP complements a scientist's hypothesis-test derivation of such algorithms. The featured solution consists of a standard non-ADF GP that incorporates a dynamic fitness function.
BOOK CHAPTER1996: Adv. in Genetic Programming 2Daida, J.M., Hommes, J.D., Bersano-Begey, T., Ross, S.J., Vesecky, J.F., (1996)." Algorithm Discovery Using the Genetic Programming Paradigm: Extracting Low-Contrast Curvilinear Features from SAR Images of Arctic Ice". Advances in Genetic Programming II , P. Angeline and K. Kinnear, Jr. (ed.) Cambridge: The MIT Press. Cited by 38 other publications.Abstract:This chapter discusses the application of genetic programming (GP) to image analysis problems in geoscience and remote sensing and describes how a GP can be adapted for processing large data sets (in our case, 1024 x 1024 pixel images plus texture channels). The featured problem is one that has not been adequately solved for this type of imagery. We describe the placement of GP in the overall scheme of algorithm discovery in geoscience image analysis and describe how GP complements a scientist's hypothesis-test derivation of such algorithms. The featured solution consists of a standard non-ADF GP that incorporates a dynamic fitness function.
I wrote chapters in 4 scientific books (left) and was invited to give a few scientific talks including the one below: Invited Talk - Location: ISI Foundation, Torino, ITALY, on Nov 27, 2009
Invited Talk - Location: ISI Foundation, Torino, ITALY, on Nov 27, 2009
Title: Automated Science, Microfluidics and Evolutionary Computation,
by Dr. Tom Bersano, University of Michigan, USASummary:The first part of this talk will cover a topic that should be of particular interest to this audience, as I will discuss Evolutionary Computation, which is a problem-solving technique that is applicable (and has been applied to) all sorts of problems, from co-evolution of interesting behaviors and strategies between different groups in simulated environments to optimization of structural designs or predictions of events or outcomes from limited data, and even to the creative computer-invention of electronic circuits that equal or surpass patented and published human designs.
I will start by introducing the general concept and giving a few salient examples from others in the field, and then briefly mention some of my own published work in improving this technique and applying it to areas like image processing or automated data mining and data analysis of satellite images, and to the design of new cellular responses and behaviors by insertion of computer-designed DNA sequences in living cells.
In the second part of this talk, I will introduce the idea of Automated Science, relating my increasing interest and progress in improving biological sciences and microfluidics to make them as accessible, reprogrammable and fast-growing as electronic devices and computers have become.
I will start by briefly introducing the main innovations responsible for the recent trend of rapid progression in biological sciences, which has made huge leaps in efficiency and throughput in its move from beakers to microarrays and is now making the next leap from microarrays to technologies like high-speed microfluidic droplet sorting.
I will then explain why unfortunately there are still key areas of research, particularly in cell biology and microfluidics itself, that simply can't be automated and sped-up this way, particularly ones involving live cells that must grow and organize on substrates over several days or that require novel experimental designs.
Finally, I will discuss my progress in completely automating and improving such areas, both by using organizing design principles such as standardization, and by designing and implementing my own working programmable, all-purpose and fully-automated lab-on-a-chip and cell-culture device.Bio:Tom Bersano is a post-doctoral researcher at the university of Michigan, currently working in the Electrical and Computer Science (EECS) Engineering Department and BioMedical Engineering (BME) Department. He obtained his PhD in EECS in 2003 from the University of Michigan under the direction of Dr. John Holland, the inventor of Genetic Algorithms. His PhD thesis was titled "Computer-evolution of gene circuits for cell-embedded computation, biotechnology, and as a model for evolutionary computation".
He holds several other Masters in Biochemistry, Biomedical Engineering, Complex Systems, Aerospace Engineering, Computer Science, and other fields.
He has published and conducted research in various areas including microfluidics, artificial intelligence, cell biology, robotics, image processing, bioinformatics, biomedical engineering and systems biology. His current research is focused on the area of "automated science" in particular with applications to microfluidics and cell manipulation, high-throughput microbiology, computer science and both theoretical and applied artificial intelligence. - 2009 (Book)
- 8 BioTechnology Conferences
- 2012 Cancer

 Cancer Research: Therapeutic Targeting of Cancer Stem CellsMalik,F; Ithimakin,S; Day,K,Ignatoski,K; Zen,Q; Thomas,D; Bersano,T; Dawsey,S; Hall,C; Davis,A; Quraishi,A; Tawakkol,N; Clouthier,S; Day, M; Hayes,D; Korkaya,H; Wicha,M. "HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: Implications for efficacy of adjuvant trastuzumab", Cancer Research 2012Proceedings of the 103rd AACRAbstract:Although current breast treatment guidelines limit the use of HER2 blocking agents such as trastuzumab to women whose tumors display HER2 gene amplification, recent retrospective analysis by Paik et al., have suggested that a wider group of patients may benefit from this therapy. Utilizing breast cancer cell lines, mouse xenograft models and primary human tumors and metastatic tissues, we provide a potential molecular explanation for these unanticipated clinical finding by demonstrating that HER2 is selectively expressed in the cancer stem cell (CSC) population in luminal ER+ breast cancers that do not display HER2 amplification. Furthermore, we demonstrate that the effects of trastuzumab on mouse tumor xenografts are highly dependent on the timing of administration. While the effects of trastuzumab on established tumors are limited to those with HER2 amplification, when this HER2 blocker is given immediately after tumor inoculation simulating an adjuvant setting, it significantly inhibits the growth of luminal tumors in the absence of HER2 amplification. In addition, HER2 expression is increased in luminal tumors propagated in mouse tibias as compared to primary tumors grown in mammary fat pads. Utilizing co-culture experiments with human osteocytes, we provide evidence that this regulated expression of HER2 is mediated by RANK ligand present in the bone microenvironment. The clinical relevance of these studies is demonstrated in breast cancer tissues by the co-expression of HER2 and CSC marker ALDH1 in a subset of cells which are selectively found at the invasive tumor front in luminal breast cancers. Recapitulating the mouse xenograft studies, we demonstrate increased expression of HER2 in bone metastasis compared to matched primary tumors in 8 patients with luminal breast cancer. Our results have significant clinical implications since they suggest that the benefits of adjuvant HER2 targeting therapies may extend to patients with luminal breast cancers who are currently classified as HER2-negative. Furthermore, these results support the cancer stem cell hypothesis and emphasize the clinical importance of targeting cancer stem cells in the adjuvant setting.
Cancer Research: Therapeutic Targeting of Cancer Stem CellsMalik,F; Ithimakin,S; Day,K,Ignatoski,K; Zen,Q; Thomas,D; Bersano,T; Dawsey,S; Hall,C; Davis,A; Quraishi,A; Tawakkol,N; Clouthier,S; Day, M; Hayes,D; Korkaya,H; Wicha,M. "HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: Implications for efficacy of adjuvant trastuzumab", Cancer Research 2012Proceedings of the 103rd AACRAbstract:Although current breast treatment guidelines limit the use of HER2 blocking agents such as trastuzumab to women whose tumors display HER2 gene amplification, recent retrospective analysis by Paik et al., have suggested that a wider group of patients may benefit from this therapy. Utilizing breast cancer cell lines, mouse xenograft models and primary human tumors and metastatic tissues, we provide a potential molecular explanation for these unanticipated clinical finding by demonstrating that HER2 is selectively expressed in the cancer stem cell (CSC) population in luminal ER+ breast cancers that do not display HER2 amplification. Furthermore, we demonstrate that the effects of trastuzumab on mouse tumor xenografts are highly dependent on the timing of administration. While the effects of trastuzumab on established tumors are limited to those with HER2 amplification, when this HER2 blocker is given immediately after tumor inoculation simulating an adjuvant setting, it significantly inhibits the growth of luminal tumors in the absence of HER2 amplification. In addition, HER2 expression is increased in luminal tumors propagated in mouse tibias as compared to primary tumors grown in mammary fat pads. Utilizing co-culture experiments with human osteocytes, we provide evidence that this regulated expression of HER2 is mediated by RANK ligand present in the bone microenvironment. The clinical relevance of these studies is demonstrated in breast cancer tissues by the co-expression of HER2 and CSC marker ALDH1 in a subset of cells which are selectively found at the invasive tumor front in luminal breast cancers. Recapitulating the mouse xenograft studies, we demonstrate increased expression of HER2 in bone metastasis compared to matched primary tumors in 8 patients with luminal breast cancer. Our results have significant clinical implications since they suggest that the benefits of adjuvant HER2 targeting therapies may extend to patients with luminal breast cancers who are currently classified as HER2-negative. Furthermore, these results support the cancer stem cell hypothesis and emphasize the clinical importance of targeting cancer stem cells in the adjuvant setting. - 2010 MicroCn

 Micro Total Analysis Systems 2010Ingram, P; Kim, YJ; Bersano-Begey, T; Lou, X; Asakura, A; Yoon, E. "Microfluidic assay to compare secretion vs contact based cell-cell interactions using dynamic isolation control", Proceedings of the 14th International Conference on Miniaturized Systems for Chemistry and Life Sciences. (MicroTAS: Micro Total-Analysis Systems).Abstract:We report cell-cell interaction studies utilizing a microfluidic chip capable of capturing cell pairs of different types into culture microwells and isolating the captured cells for a given amount of time using pneumatic valves. We have quantified the capture efficiency and confirmed cell viability and interaction in the microwells. We successfully distinguished interaction modes in the fabricated devices: secretion-based and contact-based. C2C12 cells, when cocultured with PC3, showed increased proliferation due to PC3 secretions. Also, it was indentified that C2C12 proliferation is increased through contact with endothelial cells by comparing proliferation in both secretion only and contact only systems, respectively.
Micro Total Analysis Systems 2010Ingram, P; Kim, YJ; Bersano-Begey, T; Lou, X; Asakura, A; Yoon, E. "Microfluidic assay to compare secretion vs contact based cell-cell interactions using dynamic isolation control", Proceedings of the 14th International Conference on Miniaturized Systems for Chemistry and Life Sciences. (MicroTAS: Micro Total-Analysis Systems).Abstract:We report cell-cell interaction studies utilizing a microfluidic chip capable of capturing cell pairs of different types into culture microwells and isolating the captured cells for a given amount of time using pneumatic valves. We have quantified the capture efficiency and confirmed cell viability and interaction in the microwells. We successfully distinguished interaction modes in the fabricated devices: secretion-based and contact-based. C2C12 cells, when cocultured with PC3, showed increased proliferation due to PC3 secretions. Also, it was indentified that C2C12 proliferation is increased through contact with endothelial cells by comparing proliferation in both secretion only and contact only systems, respectively. - 2009a MicroCn

 Micro Total Analysis Systems 2009Mosadegh, B; Kuo, CH; Tung, YC; Torisawa, YS; Bersano-Begey, T; Tavana, H; Takayama, S. " Scalable Integration of Elastomeric Components for Self-Regulating Microfluidic Devices", Proceedings of the 13th International Conference on Miniaturized Systems for Chemistry and Life Sciences. (MicroTAS: Micro Total-Analysis Systems).Abstract:One main research focus in microfluidics has been enhancing both the use and capabilities of devices through development of control systems1. Typically these systems achieve dynamic on-chip fluid regulation by utilizing computerized external actuators which directly valve and pump fluids in the device. Although these systems provide immensely versatile microfluidic control, their external control setups limit their scalability and convenience for use by non-specialists. This shortcoming has motivated efforts to design control systems that passively/automatically regulate fluid flow on-chip. Currently methods have only been able to passively regulate fluid in response to a dynamic input signal. Previously our group has shown the use of an elastomeric passive check-valve to temporally regulate the output of a fluid that is infused at a constant rate and therefore requires no dynamic input4. We demonstrated that a network of these valves could sequentially release and switch fluids in a finite step process.
Micro Total Analysis Systems 2009Mosadegh, B; Kuo, CH; Tung, YC; Torisawa, YS; Bersano-Begey, T; Tavana, H; Takayama, S. " Scalable Integration of Elastomeric Components for Self-Regulating Microfluidic Devices", Proceedings of the 13th International Conference on Miniaturized Systems for Chemistry and Life Sciences. (MicroTAS: Micro Total-Analysis Systems).Abstract:One main research focus in microfluidics has been enhancing both the use and capabilities of devices through development of control systems1. Typically these systems achieve dynamic on-chip fluid regulation by utilizing computerized external actuators which directly valve and pump fluids in the device. Although these systems provide immensely versatile microfluidic control, their external control setups limit their scalability and convenience for use by non-specialists. This shortcoming has motivated efforts to design control systems that passively/automatically regulate fluid flow on-chip. Currently methods have only been able to passively regulate fluid in response to a dynamic input signal. Previously our group has shown the use of an elastomeric passive check-valve to temporally regulate the output of a fluid that is infused at a constant rate and therefore requires no dynamic input4. We demonstrated that a network of these valves could sequentially release and switch fluids in a finite step process. - 2009b MicroCn

 Micro Total Analysis Systems 2009Kim, YJ; Bersano-Begey, T; Lou, X; Chung, J; Yoon, E. " Microfluidic array chip for analysis of pairwise cell interaction by temporal stimulation of secreted factors using chamber isolation", Proceedings of the 13th International Conference on Miniaturized Systems for Chemistry and Life Sciences. (MicroTAS: Micro Total-Analysis Systems).Abstract:We proposes a new microfluidic array chip which can place cell pairs of two different combination in each microchamber and analyze pairwise cell interaction in single isolated microchamber by applying pneumatic pressure. We have observed the effect of temporal stimulation of growth factors which can affects cell proliferation differences in our preliminary study. We also demonstrated the loading of the mixed pairs of PC3 and C2C12 cells into multiple separate microchambers and successive attachment and healthy morphology after culture.
Micro Total Analysis Systems 2009Kim, YJ; Bersano-Begey, T; Lou, X; Chung, J; Yoon, E. " Microfluidic array chip for analysis of pairwise cell interaction by temporal stimulation of secreted factors using chamber isolation", Proceedings of the 13th International Conference on Miniaturized Systems for Chemistry and Life Sciences. (MicroTAS: Micro Total-Analysis Systems).Abstract:We proposes a new microfluidic array chip which can place cell pairs of two different combination in each microchamber and analyze pairwise cell interaction in single isolated microchamber by applying pneumatic pressure. We have observed the effect of temporal stimulation of growth factors which can affects cell proliferation differences in our preliminary study. We also demonstrated the loading of the mixed pairs of PC3 and C2C12 cells into multiple separate microchambers and successive attachment and healthy morphology after culture. - 2009c MicroCn

 MicroTAS 2009 ConferenceChung, J; Bersano-Begey, T; Pienta, K; Yoon, E. " Clonal culture and chemodrug assay of heterogeneous cells (pc3 prostate carcinoma cells) using microfluidic single cell array chips "Proceedings of the 13th International Conference on Miniaturized Systems for Chemistry and Life Sciences.Abstract:We present a single cell assay microfluidic chip that can 1) load cells into a mi-crowell array at single cell resolution with a high capturing efficiency, 2) grow them into homogeneous clonal colonies without cell migration between neighbouring mi-crowells, and 3) supply cell culture media or reagents to each microwell from a per-fusion gravity flow induced by pressure difference between inlet and outlet reser-voirs. Using the fabricated chip, we successfully identified three clonal phenotypes in prostate cancer cells and observed the drug responsiveness varies in different sub-types.
MicroTAS 2009 ConferenceChung, J; Bersano-Begey, T; Pienta, K; Yoon, E. " Clonal culture and chemodrug assay of heterogeneous cells (pc3 prostate carcinoma cells) using microfluidic single cell array chips "Proceedings of the 13th International Conference on Miniaturized Systems for Chemistry and Life Sciences.Abstract:We present a single cell assay microfluidic chip that can 1) load cells into a mi-crowell array at single cell resolution with a high capturing efficiency, 2) grow them into homogeneous clonal colonies without cell migration between neighbouring mi-crowells, and 3) supply cell culture media or reagents to each microwell from a per-fusion gravity flow induced by pressure difference between inlet and outlet reser-voirs. Using the fabricated chip, we successfully identified three clonal phenotypes in prostate cancer cells and observed the drug responsiveness varies in different sub-types. - 2009 BioPhot
 Nanoscale Imaging for Biomedical Applications Conference
Nanoscale Imaging for Biomedical Applications Conference Society for Optical Engineering 2009Guo, Y; Ye, JY; Divin, C; Thomas, TP; Myc, A; Bersano-Begey, T; Baker, JR; Norris, TB. " Label-free biosensing using a photonic crystal structure in a total-internal-reflection geometry " in Nanoscale Imaging, Sensing, and Actuation for Biomedical Applications VI. Proceedings of the SPIE, Volume 7188 (2009)., pp. 71880B-71880B-12.Abstract:A novel optical biosensor using a one-dimensional photonic crystal structure in a total-internal-reflection geometry (PC-TIR) is presented and investigated for label-free biosensing applications. This simple configuration forms a micro Fabry-Perot resonator in the top layer which provides a narrow optical resonance to enable label-free, highly sensitive measurements for the presence of analytes on the sensing surface or the refractive index change of the surrounding medium in the enhanced evanescent field; and at the same time it employs an open sensing surface for real-time biomolecular binding detection. The high sensitivity of the sensor was experimentally demonstrated by bulk solvent refractive index changes, ultrathin molecular films adsorbed on the sensing surface, and real-time analytes binding, measuring both the spectral shift of the photonic crystal resonance and the change of the intensity ratio in a differential reflectance measurement. Detection limits of 7×10^8 RIU for bulk solvent refractive index, 6×10^5 nm for molecular layer thickness and 24 fg/mm2 for mass density were obtained, which represent a significant improvement relative to state-of-the-art surface-plasmon-resonance (SPR)-based systems. The PC-TIR sensor is thus seen to be a promising technology platform for high sensitivity and accurate biomolecular detection.
Society for Optical Engineering 2009Guo, Y; Ye, JY; Divin, C; Thomas, TP; Myc, A; Bersano-Begey, T; Baker, JR; Norris, TB. " Label-free biosensing using a photonic crystal structure in a total-internal-reflection geometry " in Nanoscale Imaging, Sensing, and Actuation for Biomedical Applications VI. Proceedings of the SPIE, Volume 7188 (2009)., pp. 71880B-71880B-12.Abstract:A novel optical biosensor using a one-dimensional photonic crystal structure in a total-internal-reflection geometry (PC-TIR) is presented and investigated for label-free biosensing applications. This simple configuration forms a micro Fabry-Perot resonator in the top layer which provides a narrow optical resonance to enable label-free, highly sensitive measurements for the presence of analytes on the sensing surface or the refractive index change of the surrounding medium in the enhanced evanescent field; and at the same time it employs an open sensing surface for real-time biomolecular binding detection. The high sensitivity of the sensor was experimentally demonstrated by bulk solvent refractive index changes, ultrathin molecular films adsorbed on the sensing surface, and real-time analytes binding, measuring both the spectral shift of the photonic crystal resonance and the change of the intensity ratio in a differential reflectance measurement. Detection limits of 7×10^8 RIU for bulk solvent refractive index, 6×10^5 nm for molecular layer thickness and 24 fg/mm2 for mass density were obtained, which represent a significant improvement relative to state-of-the-art surface-plasmon-resonance (SPR)-based systems. The PC-TIR sensor is thus seen to be a promising technology platform for high sensitivity and accurate biomolecular detection. - 1996a GeoConf
 Geoscience and Remote Sensing SymposiumCited by 28Journals
Geoscience and Remote Sensing SymposiumCited by 28Journals Geoscience and Remote Sensing 1996Daida, J.M., Bersano-Begey, T.F., Ross, S.J., Vesecky, J.F. (1996) Evolving feature-extraction algorithms: adapting genetic programming for image analysis in geoscience and remote sensing. Proceedings of the International Geoscience and Remote Sensing Symposium, IGARSS 1996, VOL 4, pages 2077-2079Abstract:Discusses a relatively new procedure in the computer-assisted design of pattern-extraction algorithms. The procedure involves the adaptation of genetic programming, a recent technique that has been used for automatic programming, for image processing and analysis. This paper summarizes several of the measures the authors have taken to develop two prototype systems that help a user to design pattern-extraction algorithms
Geoscience and Remote Sensing 1996Daida, J.M., Bersano-Begey, T.F., Ross, S.J., Vesecky, J.F. (1996) Evolving feature-extraction algorithms: adapting genetic programming for image analysis in geoscience and remote sensing. Proceedings of the International Geoscience and Remote Sensing Symposium, IGARSS 1996, VOL 4, pages 2077-2079Abstract:Discusses a relatively new procedure in the computer-assisted design of pattern-extraction algorithms. The procedure involves the adaptation of genetic programming, a recent technique that has been used for automatic programming, for image processing and analysis. This paper summarizes several of the measures the authors have taken to develop two prototype systems that help a user to design pattern-extraction algorithms - 1996b GeoConf
 Geoscience and Remote Sensing SymposiumCited by 19Journals
Geoscience and Remote Sensing SymposiumCited by 19Journals Geoscience and Remote Sensing 1996Daida, J. M. Onstott, R. G. Bersano-Begey, T. F. Ross, S. J. Vesecky, J. F. (1996) "Ice Roughness Classification and ERS SAR Imagery of Arctic Sea Ice: Evaluation of Feature-Extraction Algorithms by Genetic Programming". Proceedings of the International Geoscience and Remote Sensing Symposium, IGARSS 1996, VOL 3, pages 1520-1522Abstract:This paper describes a validation of accuracy associated with a recent algorithm that has been designed to extract ridge and rubble features from multiyear ice. Results show that the algorithm performs well with low-resolution ERS SAR data products.
Geoscience and Remote Sensing 1996Daida, J. M. Onstott, R. G. Bersano-Begey, T. F. Ross, S. J. Vesecky, J. F. (1996) "Ice Roughness Classification and ERS SAR Imagery of Arctic Sea Ice: Evaluation of Feature-Extraction Algorithms by Genetic Programming". Proceedings of the International Geoscience and Remote Sensing Symposium, IGARSS 1996, VOL 3, pages 1520-1522Abstract:This paper describes a validation of accuracy associated with a recent algorithm that has been designed to extract ridge and rubble features from multiyear ice. Results show that the algorithm performs well with low-resolution ERS SAR data products.



 My published articles in Engineering and Biotechnology conferences.There are several aspects to a Research project, including biological results that fit well within a scientific journal, and details on engineering, design and fabrication that might be more appropriate for engineering conferences. I published some of these results in international conferences for Cancer Research, Micro- Total Analysis Systems, Geoscience and Remote Sensing Symposiums, and Optical Engineering Conferences.
My published articles in Engineering and Biotechnology conferences.There are several aspects to a Research project, including biological results that fit well within a scientific journal, and details on engineering, design and fabrication that might be more appropriate for engineering conferences. I published some of these results in international conferences for Cancer Research, Micro- Total Analysis Systems, Geoscience and Remote Sensing Symposiums, and Optical Engineering Conferences.



- 2012 Cancer
- 7 Computer Science Articles
- 2002 AiConf
 Genetic & Evolutionary Programming Conference
Genetic & Evolutionary Programming Conference Genetic and Evolutionary Computation Conference 2002Bersano-Begey, T.F., (2002). Cellular Programming and Bioware: Computer Evolution of Gene Circuits to Re-Program Living Cells. Proceeding of the Evolutionary Computation in the Industry track in the Genetic and Evolutionary Computation Conference 2002, New York, NY.Abstract:The current state of the art in biotechnology allows us to engineer gene circuits in real living cells that perform predictable switching and dynamic behavior. The design of such circuits has vast potential for the fabrication of biological devices with a new range of applications, yet this field still lacks a developed discipline of rational circuit engineering. However, several aspects of the underlying biology make the design of complex gene circuits not addressable by standard circuit design and engineering techniques. This paper discusses the main features of this new field and shows how the automated design of complex gene-circuits that are comparable or more complex than ones designed by humans is possible through the use of a modified Classifier System (CS) and Genetic Algorithm (GA). In particular, this paper shows all the steps involved in the evolution of a gene circuit to program bacteria to target and swim toward enemy cells and destroy them, and the usefulness of a simulation environment to test gene circuit behaviors. A language encoding gene circuits is developed and its translation into actual DNA sequences and biological manipulations and implementation is discussed.
Genetic and Evolutionary Computation Conference 2002Bersano-Begey, T.F., (2002). Cellular Programming and Bioware: Computer Evolution of Gene Circuits to Re-Program Living Cells. Proceeding of the Evolutionary Computation in the Industry track in the Genetic and Evolutionary Computation Conference 2002, New York, NY.Abstract:The current state of the art in biotechnology allows us to engineer gene circuits in real living cells that perform predictable switching and dynamic behavior. The design of such circuits has vast potential for the fabrication of biological devices with a new range of applications, yet this field still lacks a developed discipline of rational circuit engineering. However, several aspects of the underlying biology make the design of complex gene circuits not addressable by standard circuit design and engineering techniques. This paper discusses the main features of this new field and shows how the automated design of complex gene-circuits that are comparable or more complex than ones designed by humans is possible through the use of a modified Classifier System (CS) and Genetic Algorithm (GA). In particular, this paper shows all the steps involved in the evolution of a gene circuit to program bacteria to target and swim toward enemy cells and destroy them, and the usefulness of a simulation environment to test gene circuit behaviors. A language encoding gene circuits is developed and its translation into actual DNA sequences and biological manipulations and implementation is discussed. - 1997a AiConf
 Genetic Programming ConferenceCited by 11Journals
Genetic Programming ConferenceCited by 11Journals Genetic Programming 1997Bersano-Begey, T.F., Daida, J.M., (1997). A Discussion on Generality and Robustness and A Framework for Fitness Set Construction in Genetic Programming to Promote Robustness. Proceedings of the Genetic Programming 1997 conference (lbp), Stanford University, Cambridge, MA: The MIT Press.Abstract:A significant problem in Genetic Programming consists in ensuring the robustness or generality of the evolved code (its ability to work correctly on never before seen data). We examine approaches attempted so far, and propose a different solution, based on multiple training sets (to discriminate between possible interpretations), augmentation and refinement, which improves on both task specification and distribution of fitness. We then present some preliminary results on its application to a fairly canonical GP problem (wall-following behavior) that has been previously shown to be very brittle. Finally, we discuss issues in using additional information about the generality of individuals.
Genetic Programming 1997Bersano-Begey, T.F., Daida, J.M., (1997). A Discussion on Generality and Robustness and A Framework for Fitness Set Construction in Genetic Programming to Promote Robustness. Proceedings of the Genetic Programming 1997 conference (lbp), Stanford University, Cambridge, MA: The MIT Press.Abstract:A significant problem in Genetic Programming consists in ensuring the robustness or generality of the evolved code (its ability to work correctly on never before seen data). We examine approaches attempted so far, and propose a different solution, based on multiple training sets (to discriminate between possible interpretations), augmentation and refinement, which improves on both task specification and distribution of fitness. We then present some preliminary results on its application to a fairly canonical GP problem (wall-following behavior) that has been previously shown to be very brittle. Finally, we discuss issues in using additional information about the generality of individuals. - 1997b AiConf
 Genetic Programming ConferenceCited by 07Journals
Genetic Programming ConferenceCited by 07Journals Genetic Programming 1997Bersano-Begey, T.F., (1997) Controlling Exploration, Diversity and Escaping Local Optima in GP: Adapting Weights of Training Sets to Model Resource Consumption. Proceedings of the Genetic Programming 1997 conference (lbp), Stanford University, Cambridge, MA: The MIT Press.Abstract:A common problem in evolutionary computation, and in particular in GA and GP, is the loss of diversity due both to `lock-in' and early convergence of a population to `deceptive' high scoring partial solutions. As the run proceeds, such individuals constitute a larger and larger segment of the population, eliminate diversity and stop the progress of the run. This paper will present a method for the automatic detection of such lock-in and impasse, as well as explore a fitness function which uses this information to reward diversity and innovation and increasingly penalize the locked-in individuals. This method can be applied to essentially any problem, and can be successful in preventing a run from locking-in on an easily obtainable local optimum and in preserving diversity. We present an analysis, discussion, and preliminary results on its application to the boolean 11-multiplexer problem.
Genetic Programming 1997Bersano-Begey, T.F., (1997) Controlling Exploration, Diversity and Escaping Local Optima in GP: Adapting Weights of Training Sets to Model Resource Consumption. Proceedings of the Genetic Programming 1997 conference (lbp), Stanford University, Cambridge, MA: The MIT Press.Abstract:A common problem in evolutionary computation, and in particular in GA and GP, is the loss of diversity due both to `lock-in' and early convergence of a population to `deceptive' high scoring partial solutions. As the run proceeds, such individuals constitute a larger and larger segment of the population, eliminate diversity and stop the progress of the run. This paper will present a method for the automatic detection of such lock-in and impasse, as well as explore a fitness function which uses this information to reward diversity and innovation and increasingly penalize the locked-in individuals. This method can be applied to essentially any problem, and can be successful in preventing a run from locking-in on an easily obtainable local optimum and in preserving diversity. We present an analysis, discussion, and preliminary results on its application to the boolean 11-multiplexer problem. - 1997c AiConf
 IEEE Evolutionary Computation ConferenceCited by 06Journals
IEEE Evolutionary Computation ConferenceCited by 06Journals IEEE - Evolutionary Computation 1997Bersano-Begey, T.F., Daida, J.M., Vesecky, J.F., Ludwig, F.L., (1997). A Java Collaborative Interface for Genetic Programming Applications: Image Analysis for Scientific Inquiry . Proceedings of the 1997 IEEE International Conference of Evolutionary Computation, IEEE Press.Abstract:This paper discusses several key issues involved in designing and using a Java collaborative interface for genetic programming applications over the World Wide Web. We present our implementation that has been used in a new system that assists scientists in classifying and extracting novel features in remotely sensed satellite imagery. This paper also identifies issues in developing a class library that facilitates rapid prototyping of such collaborative graphical user interfaces for genetic programming, and suggests how other researchers could benefit from them.
IEEE - Evolutionary Computation 1997Bersano-Begey, T.F., Daida, J.M., Vesecky, J.F., Ludwig, F.L., (1997). A Java Collaborative Interface for Genetic Programming Applications: Image Analysis for Scientific Inquiry . Proceedings of the 1997 IEEE International Conference of Evolutionary Computation, IEEE Press.Abstract:This paper discusses several key issues involved in designing and using a Java collaborative interface for genetic programming applications over the World Wide Web. We present our implementation that has been used in a new system that assists scientists in classifying and extracting novel features in remotely sensed satellite imagery. This paper also identifies issues in developing a class library that facilitates rapid prototyping of such collaborative graphical user interfaces for genetic programming, and suggests how other researchers could benefit from them. - 1997d AiConf
 Evolutionary Programming ConferenceCited by 12Journals
Evolutionary Programming ConferenceCited by 12Journals Evolutionary Programming 1997Polito, J., Daida, J.M., Bersano-Begey, T.F., (1997). Musica ex Machina: Composing 16th-Century Counter-point with Genetic Programming and Symbiosis . Proceedings of the VI International Conference of Evolutionary Programming (EP97), Angeline et al. (Eds), Springer Press.Abstract:GPmuse is software which explores one connection between computation and creativity using a symbiosis-inspired genetic programming paradigm in which distinct agents collaborate to produce 16th-century counterpoint.
Evolutionary Programming 1997Polito, J., Daida, J.M., Bersano-Begey, T.F., (1997). Musica ex Machina: Composing 16th-Century Counter-point with Genetic Programming and Symbiosis . Proceedings of the VI International Conference of Evolutionary Programming (EP97), Angeline et al. (Eds), Springer Press.Abstract:GPmuse is software which explores one connection between computation and creativity using a symbiosis-inspired genetic programming paradigm in which distinct agents collaborate to produce 16th-century counterpoint. - 1996a AiConf
 Genetic Programming ConferenceCited by 14Journals
Genetic Programming ConferenceCited by 14Journals Genetic Programming 1996Ross, S.J., Daida, J.M., Doan, C.M., Bersano-Begey, T.F., McClain, J.J, (1996). Variations in evolution of subsumption architectures using genetic programming: The Wall Following Robot Revisited . Proceedings of the 1996 Genetic Programming Conference, July 28-31, 1996, Stanford University, Cambridge, MA: The MIT Press.Abstract:The wall following robot is examined as a potential benchmark problem for applications of genetic programming (GP) to emergent robotic behavior. This paper describes experiments that were performed to characterize the performance, solution space, search space, and robustness using GP with and without automatically defined functions (ADFs). GP with ADFs was unable to significantly outperform GP without ADFs on this problem. A suboptimal modality was discovered across all four program architectures. Many of the optimal solutions that were discovered tended to limit the number of sensors used for wall-following behavior; some used as few as three sensors. Tests for robustness indicate a "handedness" to the evolved solutions, which does seem to contribute to solution brittleness.
Genetic Programming 1996Ross, S.J., Daida, J.M., Doan, C.M., Bersano-Begey, T.F., McClain, J.J, (1996). Variations in evolution of subsumption architectures using genetic programming: The Wall Following Robot Revisited . Proceedings of the 1996 Genetic Programming Conference, July 28-31, 1996, Stanford University, Cambridge, MA: The MIT Press.Abstract:The wall following robot is examined as a potential benchmark problem for applications of genetic programming (GP) to emergent robotic behavior. This paper describes experiments that were performed to characterize the performance, solution space, search space, and robustness using GP with and without automatically defined functions (ADFs). GP with ADFs was unable to significantly outperform GP without ADFs on this problem. A suboptimal modality was discovered across all four program architectures. Many of the optimal solutions that were discovered tended to limit the number of sensors used for wall-following behavior; some used as few as three sensors. Tests for robustness indicate a "handedness" to the evolved solutions, which does seem to contribute to solution brittleness. - 1996b AiConf
 Genetic Programming ConferenceCited by 44Journals
Genetic Programming ConferenceCited by 44Journals Genetic Programming 1996Daida, J.M., Bersano-Begey, T.F., Ross, S.J, Vesecky J.F., (1996). Computer-Assisted Design of Image Analysis Algorithms: Dynamic and Static Fitness Evaluations in a Scaffolded Environment . Proceedings of the 1996 Genetic Programming Conference, July 28-31, 1996, Stanford University, Cambridge, MA: The MIT Press.Abstract:This paper discusses several issues in applying genetic programming to image classification problems in geoscience and remote sensing. In particular, this paper examines the role in using dynamic and static fitness evaluation functions. This paper also examines a few of the aspects in human-computer interactions that facilitate computer-assisted learning and problem solving (i.e., scaffolding) for our system. We describe a possible means for visualizing and summarizing a solution space without having to resort to an exhaustive search of individuals.
Genetic Programming 1996Daida, J.M., Bersano-Begey, T.F., Ross, S.J, Vesecky J.F., (1996). Computer-Assisted Design of Image Analysis Algorithms: Dynamic and Static Fitness Evaluations in a Scaffolded Environment . Proceedings of the 1996 Genetic Programming Conference, July 28-31, 1996, Stanford University, Cambridge, MA: The MIT Press.Abstract:This paper discusses several issues in applying genetic programming to image classification problems in geoscience and remote sensing. In particular, this paper examines the role in using dynamic and static fitness evaluation functions. This paper also examines a few of the aspects in human-computer interactions that facilitate computer-assisted learning and problem solving (i.e., scaffolding) for our system. We describe a possible means for visualizing and summarizing a solution space without having to resort to an exhaustive search of individuals.
My published articles in Computer Science, Machine Learning and Artificial Intelligence Conferences.In computer science, differently from fields like the life sciences, conferences can often be more influential, produce more citations, and be more instrumental in advancing the field than journals.
These publications are my earlier work on aritficial intelligence and in particularly in both theory and applications of evolutionary computation, a method to do complex problem-solving by simulating survival of the fittest and evolution of solutions, designs and automatically generated algorithms.



- 2002 AiConf
- 2010 CCG

 Part (of this grant) that I wrote:90%Budget: ~$100,000 (1 year)Status:Successfully FundedCollaborators/PIs:YoonTitle:"A High-Throughput Low-Cost Portable Technology for Single-Cell and Cell-Cell Interaction Screening"Goal:Research to advance combinatorial testing of cell-cell interactions
Part (of this grant) that I wrote:90%Budget: ~$100,000 (1 year)Status:Successfully FundedCollaborators/PIs:YoonTitle:"A High-Throughput Low-Cost Portable Technology for Single-Cell and Cell-Cell Interaction Screening"Goal:Research to advance combinatorial testing of cell-cell interactions - 2009 NIH

- 2009 IMAT

 Part (of this grant) that I wrote:70%Budget: ~$500,000 (R21)Collaborators/PIs:Pienta, YoonTitle:(protected)Goal:Detect and isolate hard-to-detect cancer stem cells based on their growth dynamic.
Part (of this grant) that I wrote:70%Budget: ~$500,000 (R21)Collaborators/PIs:Pienta, YoonTitle:(protected)Goal:Detect and isolate hard-to-detect cancer stem cells based on their growth dynamic. - 2009 NIST

 Part (of this grant) that I wrote:50%Budget: ~$1 millionCollaborators/PIs:Kopelmann,YoonTitle:High-throughput Microscope-free Real-time Cellular Monitoring Devices: Combining Nanoprobes and MicrofluidicsGoal:Using nanoprobes in microdevices for new high-throughput cell assays
Part (of this grant) that I wrote:50%Budget: ~$1 millionCollaborators/PIs:Kopelmann,YoonTitle:High-throughput Microscope-free Real-time Cellular Monitoring Devices: Combining Nanoprobes and MicrofluidicsGoal:Using nanoprobes in microdevices for new high-throughput cell assays - 2009 NCI

- 2009 SBIR

- 2009 CCNE

- 2009 NCI

- 2008 NSF

- 2000 DARPA

 Part (of this grant) that I wrote:5%Budget: ~$10 MillionsCollaborators/PIs:Nori and 17 othersTitle:"Fundamental Research at the interface between biology, computing, and micro-technology: networks and computing in biological and artificial systems"Goal:(protected)
Part (of this grant) that I wrote:5%Budget: ~$10 MillionsCollaborators/PIs:Nori and 17 othersTitle:"Fundamental Research at the interface between biology, computing, and micro-technology: networks and computing in biological and artificial systems"Goal:(protected) - Develop micro- and nanointuition. (lectures/exams on scaling laws, surfaces, and specific examples)
- Learn basic methods of micro- and nanofabrication, especially the methods most useful for biological applications. (lectures on photolithography, soft lithography, self-assembly)
- Familiarization with micro- and nanotechnology literature and applications. (presentation, literature research, proposals)
- Learn how to design, evaluate, and analyze micro- and nanodevices. (lectures, discussions, presentations, proposals)
- Sharpen critical thinking and analysis (discussions, presentations, proposals)
- Practice effective communication of ideas (presentations, proposals)
- Cultivate innovative thinking (proposals, discussions)
- Learn how to learn new fields (literature research, presentations, proposals)
- Learn teamwork (proposal projects)
- Learn what is available/goinghere at Michigan (field trips, guest lectures)
- Managing potentially overwhelming input (even before e-mail there was too much going on)
- Selective Learning (so how should one decide what to remember?)
- Knowledge integration (and how does one do that without destroying what's already there?)
- Control mechanisms (need different ways to deal with important vs. unimportant, relevant vs. irrelevant)
- PDP
- Classifier systems and genetic algorithms
- Soar and Unified Theories of Cognition
- EPIC
- ACT-R and rational analysis
- Symbolic vs. subsymbolic approaches
- Alternative views DOMAINS
- HCI
- Case-based reasoning
- Deductive reasoning
- Skill acquisition
- Language acquisition
- Semantic memory
- Categorization
- Attention
- Working Memory
- Explicit learning and memory
- Vision
- Neural organization FINAL DISCUSSION AND PROJECTS PRESENTATIONS
- In what sense, if any, is there a need for theory in morality?
- How are we to understand the meaning of moral terms?
- Are moral judgments capable of truth and falsity?
- In what sense, if any, can moral claims be objective?
- How is morality related to relationality?
- What is the relation of "ought" to "is"?
- And, why be moral? Midterm and final examinations; a term paper.
- read, discuss, evaluate a number of models from a variety of disciplines.
- Modify and run experiments with exisiting models.
- Design, implement, run, write-up results from their own models. The course will cover all aspects of the modeling process itself, from model design through implementation to analyzing, documenting and communicating results.
- 1995
- 1996
- 1997
- 1998
- 1999
- 2000
- 2001
- 2002
- 2003
- 2004
- 2005
- 2006
- 2007
- 2008
- 2009
- 2010
- 2011
 UG Researcher, UM AOSS Dept.
UG Researcher, UM AOSS Dept.

 Grad Student Researcher, UM AI Lab
Grad Student Researcher, UM AI Lab
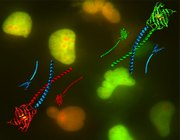 Research Lab Rotation (Gene Regulation Lab), BioChemistry Dept.
Research Lab Rotation (Gene Regulation Lab), BioChemistry Dept. PhD Research, UM AI Lab
PhD Research, UM AI Lab
 Visiting Scientist, Riken Institute (Tokyo, Japan) and UM Physics Lab
Visiting Scientist, Riken Institute (Tokyo, Japan) and UM Physics Lab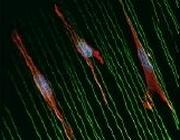

 PostDoc Researcher, UM BioMed Lab
PostDoc Researcher, UM BioMed Lab PostDoc, UM Microsystems Lab
PostDoc, UM Microsystems Lab
 Researcher, UM Oncology Lab
Researcher, UM Oncology Lab Internship, Pfizer (Parke-Davis)
Internship, Pfizer (Parke-Davis) My Position: InternshipEmployment Year: 1995
My Position: InternshipEmployment Year: 1995
Location: Ann Arbor,MI,USA.Map Street View
Company:Parke-Davis Pharmaceutical Research (now Pfizer1) was America's oldest and largest drug maker.Job Description - Area of work:There is a big component of data management to pharmaceutical companies. It takes many years and hundreds of millions of dollars to get new drugs approved and to the market, and the company must prove that experimental data has not been compromised in any way, or risk facing great added costs.Job Description - My Contributions:I was in charge of supervising the servers containing this valuable information. To protect and support years of new drug development data I developed new programs, applications and scripts for servers kept locked beyond 3 levels of security doors, and helped migration from older systems (VAX) to more recent ones (Windows). I also developed automatic back-up scheme and supervised data back-ups and restores for this data and my scripts controlled all the password accounts and password changes to this data, from ordinary workers to executives.

 Senior Engineer at General Inspection Inc.
Senior Engineer at General Inspection Inc.

 My Position: Senior EngineerEmployment Year: 1998-2000
My Position: Senior EngineerEmployment Year: 1998-2000
Company:General Inspection, produces laser-sorting equipment to serve the automotive, aerospace and gauging industries.Job Description - Area of work:Cars, Airplanes, and most machines and tools need screws, bolts and other small parts, generally made by 3rd party manufacturers. The crucial part of all this is that if a screw is ever slightly defective, your car, airplane etc could break down and whoever made them will get a bad reputation or lose money by forced recalls. And despite how sophisticated your fabrication methods are, there is always a very small fraction of parts that end up with fabrication defects. So companies have to inspect their parts, and bolt makers have to inspect the millions of bolts they make for almost invisible defects.Job Description - My Contributions:I was in charge of developing and improving new high-speed laser scanning systems to find and remove these 'needle in a haystack' defective parts. Some of these defects could only be found by a combination of laser, ultrasound, and eddy currents so I had to program and integrate these raw sensor data into a coherent laser gauging technology and software capable to analyze, measure, and sort parts with variable specifications. In addition, in order to achieve high-speed sorting of 300 parts per minute, I had to devise ways to compensate for variable speed/ bouncing/friction, effects which would otherwise disrupt the instrument's precision and render readings useless. And when looking at high precision, everything is more difficult: even small changes in temperature and environment would make the laser signal fluctuate and become unreliable, so I had to build in calibrations, interpolations high speeds (~300 parts per minute). I then built-in methods for automated statistical data collection and automatic shape recognition/analysis/fitting and measurement of relevant features, and for 3D reconstruction and display.
 SoftwareEngineer at Netarx.com
SoftwareEngineer at Netarx.com My Position: Software EngineerEmployment Year: 2001
My Position: Software EngineerEmployment Year: 2001
Location: Auburn Hills, MI, USA
Company:Netarx, a company that provides Managed IT Services.Job Description - Area of work:Larger companies with their own internal network need to keep track of all their servers, detect security threats, deploy updates and perform upgrades and repairs. One solution is to pay people to physically go around and check for any problem and maintain all these infrastructure components. But an arguably better solution is to automate and centralize this process, so that each component communicates if there are any problems that might later lead to crashes or security attacks and can be updated remotely, and technicians are only deployed when absolutely needed and knowing exactly what to replace.Job Description - My Contributions:As a Software Engineer in the Research and Development department, I worked on integrating all the incoming information from monitored network components into a coherent situational analysis to predict or deduce possible causes for any network problem, and to generate automatic notifications to support and service people. I then developed a web-based tool that allowed corporate partners to monitor and navigate through a dynamic visual representation of their entire network in which any network congestion, security breach, or problem is displayed in real time.
 Senior Engineer at Lesynski.com
Senior Engineer at Lesynski.com My Position: Senior EngineerEmployment Year: 2003
My Position: Senior EngineerEmployment Year: 2003
Location: Seattle, WA,USA.Map Street View
Company:Leszynski Group, Inc., developed several Pen-Based technologies and APIs for Microsoft.Job Description - Area of work:Several years before the iPhone, the iPad, the Nintendo-DS and other touch and pen-based devices, the technology for these new types of interfaces was still being developed and the main new platform to allow touch and pen input was the TabletPC (a touchscreen PC laptop). That's what I had to work with at the time.Job Description - My Contributions:My focus was on important applications that only become possible by combining artificial intelligence and pen-based input. For example, while inspecting and repairing military vehicles and planes, it can be important to look up the blueprint schematics of some part, but often people could sketch the part but not know its exact name and model. Before my work there was no way to search blueprints in a database based on partial sketches, so I devised new shape recognition algorithms and new conceptual and visual pattern-matching AI to perform that task.
Additionally I developed a biometric recognition algorithm that extracted speed and pressure information from signatures written with a tablet PC pen and used them to perform authentication and cryptography. While the looks of a signature can be forged, the general pattern of pressure and acceleration as it is written is almost impossible to guess and reproduce.
Then, through a collaboration with Microsoft, I developed a shape-recognition SDK for tablet-PCs and other pen-based applications that used artificial intelligence and complex pattern recognition to interpret and correct hand-drawn diagrams and sketches over a variety of applications and domains. This new technology was featured in multiple keynote talks by Bill Gates and other Microsoft Executives, and allowed applications like search of part databases from partial rough sketches and animating sketches into physical simulations (in collaboration with MIT) automatically.
 My Position: Science/Technology ConsultantEmployment Year: 2006-2011
My Position: Science/Technology ConsultantEmployment Year: 2006-2011
Company Type: Independent ContractorJob Description:I Developed solutions to scientific problems for industrial companies, on a project basis. I have found working solutions to problems that internal engineers had tried to solve and determined to be 'unsolvable', such as noise-free Hex and trilobe laser reconstructions.
My published articles in scientific and biotechnology journals.I have published in several high-impact scientific journals including "Science" and "Nature Physics", and in a variety of fields in journals including "Integrative Biology", "Biomaterials", "Current Opinions in Medicinal Chemistry", "Biomedical Microdevices", and "Lab on a Chip", through various collaborations and research projects.

 Note:Move the pointer over one of the tabs above to switch between my articles in Journals, Books, Biotechnology Conference, and Computer Science Conferences.My Research Skills: 24 Technologies and Techniques I MasteredComputational Technology:Artificial IntelligenceSimulation and modelingSoftware EngineeringImage ProcessingAutomation and ScriptingGenetic ProgrammingLanguages:English, ItalianSpanish, FrenchProgramming: C#, C++, Java, Perl, Javascript, CSS3, HTML5, PHP...Clean Room and Microfabrication:Microprocessors design and fabricationMEMS Micro-Electro-Mechanical SystemsNano-Technology and RoboticsSoft-Lithography and MicroFluidicsMolecular and Cell Biology:Mammalian Cell and Tissue CultureTransfection and Viral VectorsPrimary and Stem Cell CulturesCo-Culture, Single-Cell MeasurementsGenetic Engineering:Bacterial TransformationRecombinant DNA Sub-CloningPolymerase Chain Reaction MethodsChimeric Protein Design&SynthesisBiochemical and Lab Technology:Immuno-Fluorescent MicroscopyPurification of DNA and ProteinsElectrophoresis and Blots (Western, etc)DNA SynthesisMy Grants: 10 Funding/Research Proposals I wrote for the UniversityGrants and Funding Proposals I wrote for the University of Michigan.Writing grants is an important part of doing research and paying for equipment, reagents and especially the time of scientists, engineers and students that will actually do much of the work. I wrote grants for very different funding agencies, some focused on science, some on technology advancement, and others even connected to entrepeneurship and developing new businesses based on new technology, and I have learned a lot about this important task in the process.
Note:Move the pointer over one of the tabs above to switch between my articles in Journals, Books, Biotechnology Conference, and Computer Science Conferences.My Research Skills: 24 Technologies and Techniques I MasteredComputational Technology:Artificial IntelligenceSimulation and modelingSoftware EngineeringImage ProcessingAutomation and ScriptingGenetic ProgrammingLanguages:English, ItalianSpanish, FrenchProgramming: C#, C++, Java, Perl, Javascript, CSS3, HTML5, PHP...Clean Room and Microfabrication:Microprocessors design and fabricationMEMS Micro-Electro-Mechanical SystemsNano-Technology and RoboticsSoft-Lithography and MicroFluidicsMolecular and Cell Biology:Mammalian Cell and Tissue CultureTransfection and Viral VectorsPrimary and Stem Cell CulturesCo-Culture, Single-Cell MeasurementsGenetic Engineering:Bacterial TransformationRecombinant DNA Sub-CloningPolymerase Chain Reaction MethodsChimeric Protein Design&SynthesisBiochemical and Lab Technology:Immuno-Fluorescent MicroscopyPurification of DNA and ProteinsElectrophoresis and Blots (Western, etc)DNA SynthesisMy Grants: 10 Funding/Research Proposals I wrote for the UniversityGrants and Funding Proposals I wrote for the University of Michigan.Writing grants is an important part of doing research and paying for equipment, reagents and especially the time of scientists, engineers and students that will actually do much of the work. I wrote grants for very different funding agencies, some focused on science, some on technology advancement, and others even connected to entrepeneurship and developing new businesses based on new technology, and I have learned a lot about this important task in the process.


 My Coursework: 125+ Courses that I completed:Biotechnology [3]
My Coursework: 125+ Courses that I completed:Biotechnology [3] Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.MicroBiology and Immunology [9]BIO 436Immunology
Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.MicroBiology and Immunology [9]BIO 436Immunology BIO 436 - Introductory Immunology:This course is intended to introduce pre-professional and biology concentrators to the theoretical and experimental principles of immunology. Topics covered include: a detailed study of the molecules, cells, and organs that constitute the immune system; the innate and adaptive immune responses; and the role of the immune system in host defense, allergy, and organ transplantation. Topics will be illustrated with clinical case studies. Grades are based on three exams. The course is appropriate for concentrations in biology, microbiology, and cell and molecular biology.BIOCHEM 597Critical Analysis
BIO 436 - Introductory Immunology:This course is intended to introduce pre-professional and biology concentrators to the theoretical and experimental principles of immunology. Topics covered include: a detailed study of the molecules, cells, and organs that constitute the immune system; the innate and adaptive immune responses; and the role of the immune system in host defense, allergy, and organ transplantation. Topics will be illustrated with clinical case studies. Grades are based on three exams. The course is appropriate for concentrations in biology, microbiology, and cell and molecular biology.BIOCHEM 597Critical Analysis BIOCHEM 597 - Critical Analysis:A course designed to help first-year graduate students in the Ph.D. program improve their skills in reading, analyzing, discussing, and writing about the biochemical literature.MICROBIO 604Microbial. GeneticsMICROBIO 606Microbial Physiology and metabolism
BIOCHEM 597 - Critical Analysis:A course designed to help first-year graduate students in the Ph.D. program improve their skills in reading, analyzing, discussing, and writing about the biochemical literature.MICROBIO 604Microbial. GeneticsMICROBIO 606Microbial Physiology and metabolism MICROBIO 606 - Microbial Physiology and Metabolism:This module focuses on the bacterial cell as evolution's most successful biological product in terms of growth, survival, and niche colonization. Topics include the metabolism and physiology of growth as well as selected environmental response strategies, such as directed motility, resistance, mechanisms, regulation of genome expression above the operon level, and differentiation into non-growth states.MICROBIO 553Cancer Cell Biology
MICROBIO 606 - Microbial Physiology and Metabolism:This module focuses on the bacterial cell as evolution's most successful biological product in terms of growth, survival, and niche colonization. Topics include the metabolism and physiology of growth as well as selected environmental response strategies, such as directed motility, resistance, mechanisms, regulation of genome expression above the operon level, and differentiation into non-growth states.MICROBIO 553Cancer Cell Biology MICROBIO 553 - Cancer Cell Biology:This course will cover a broad range of subjects relating to cancer biology. Emphasis is on the relationship between basic science and clinical aspects of cancer. Topics to be covered include carcinogenesis, cancer progression, tumor pathology, oncogenes, cellular growth control, tumor suppressor genes, oncogenic viruses, apoptosis, tumor immunology, clinical oncology, and therapeutics. The course consists of lectures by faculty in the Cancer Center who are experts on various topics.MICROBIO 615Mol. Cellular Viral Pathogenesis
MICROBIO 553 - Cancer Cell Biology:This course will cover a broad range of subjects relating to cancer biology. Emphasis is on the relationship between basic science and clinical aspects of cancer. Topics to be covered include carcinogenesis, cancer progression, tumor pathology, oncogenes, cellular growth control, tumor suppressor genes, oncogenic viruses, apoptosis, tumor immunology, clinical oncology, and therapeutics. The course consists of lectures by faculty in the Cancer Center who are experts on various topics.MICROBIO 615Mol. Cellular Viral Pathogenesis MICROBIO 615 - Mol. Cellular Viral Pathogenesis:Will focus on interactions of selected viral pathogens with cells to successfully infect, replicate and spread in hosts. Emphasis will be on how virus replication and host cellular receptors, regulatory proteins and components of the immune system determine pathogenesis and the outcome of infection. Format combines critical evaluation of primary literature with some lecture/discussions.MICROBIO 616DNA Tumor Viruses
MICROBIO 615 - Mol. Cellular Viral Pathogenesis:Will focus on interactions of selected viral pathogens with cells to successfully infect, replicate and spread in hosts. Emphasis will be on how virus replication and host cellular receptors, regulatory proteins and components of the immune system determine pathogenesis and the outcome of infection. Format combines critical evaluation of primary literature with some lecture/discussions.MICROBIO 616DNA Tumor Viruses MICROBIO 616 - DNA Tumor Viruses:This course will cover the molecular biology of small DNA viruses, with an emphasis on tumor viruses. Topics to be discussed include replication, gene expression, oncogenic transformation, and the virus-host interactions that contribute to these processes such as viral oncoprotein-tumor suppressor protein interactions. The format will be a combination of lectures by the instructor and reading and discussion of the primary literature by the class.MICROBIO 617RetroViruses
MICROBIO 616 - DNA Tumor Viruses:This course will cover the molecular biology of small DNA viruses, with an emphasis on tumor viruses. Topics to be discussed include replication, gene expression, oncogenic transformation, and the virus-host interactions that contribute to these processes such as viral oncoprotein-tumor suppressor protein interactions. The format will be a combination of lectures by the instructor and reading and discussion of the primary literature by the class.MICROBIO 617RetroViruses MICROBIO 617 - RetroViruses:This course will consist of lectures and class discussion of primary literature and other materials about retroviruses. Students will be required to read assigned materials and display critical understanding of them, and to participate in exercises intended to strengthen their skills in the preparation and criticism of scientific manuscripts. Materials covered will include the following: a) The biology and replication of retroviruses, including the organization of their genomes, the functions of their genes, their replication cycles, and viral population dynamics. b) The RNA tumor virus class of retroviruses, including their role in the discovery of cellular oncogenes and how they cause animal tumors. c) Human retroviruses with a focus on HIV, including the biology of HIV, the roles of HIV accessory proteins, the evidence that HIV causes AIDS, and strategies for the treatment of HIV disease. d) The use of retrovirus derivatives as vectors for gene therapy.MICROBIO 812Graduate Seminar
MICROBIO 617 - RetroViruses:This course will consist of lectures and class discussion of primary literature and other materials about retroviruses. Students will be required to read assigned materials and display critical understanding of them, and to participate in exercises intended to strengthen their skills in the preparation and criticism of scientific manuscripts. Materials covered will include the following: a) The biology and replication of retroviruses, including the organization of their genomes, the functions of their genes, their replication cycles, and viral population dynamics. b) The RNA tumor virus class of retroviruses, including their role in the discovery of cellular oncogenes and how they cause animal tumors. c) Human retroviruses with a focus on HIV, including the biology of HIV, the roles of HIV accessory proteins, the evidence that HIV causes AIDS, and strategies for the treatment of HIV disease. d) The use of retrovirus derivatives as vectors for gene therapy.MICROBIO 812Graduate Seminar MICROBIO 812 - Graduate Seminar:An analysis of advances at the frontiers of microbiology. Every graduate student is required to enroll in this course every semester.BioMedical Engineering [8]MSE 250Fundamentals of Material Science Engineering
MICROBIO 812 - Graduate Seminar:An analysis of advances at the frontiers of microbiology. Every graduate student is required to enroll in this course every semester.BioMedical Engineering [8]MSE 250Fundamentals of Material Science Engineering MSE 250 - Fundamentals of Material Science Engineering:Properties (mechanical, thermal and electrical) of metals, polymers, ceramics, and electronic materials. Correlations of these properties with: (1) their internal structures (atomic, molecular, crystalline, micro-and macro); (2) service conditions (mechanical, thermal, chemical, electrical, magnetic and radiation); and (3) processing.NE&RS 400Elements of Nuclear EngineeringBIOMED 410BioMedical Materials
MSE 250 - Fundamentals of Material Science Engineering:Properties (mechanical, thermal and electrical) of metals, polymers, ceramics, and electronic materials. Correlations of these properties with: (1) their internal structures (atomic, molecular, crystalline, micro-and macro); (2) service conditions (mechanical, thermal, chemical, electrical, magnetic and radiation); and (3) processing.NE&RS 400Elements of Nuclear EngineeringBIOMED 410BioMedical Materials BIOMED 410 - BioMedical Materials:Biomaterials and their physiological interactions. Materials used in medicine/dentistry: metals, ceramics, polymers, composites , resorbable smart, natural materials. Material response/degradation: mechanical breakdown, corrosion, dissolution, leaching, chemical degradation, wear. Host responses: foreign body reactions, inflammation, wound healing, carcinogenicity, immunogenicity, cytotoxicty, infection, local/systemic effects.BIOMED 483MRI Magnetic Resonance Imaging
BIOMED 410 - BioMedical Materials:Biomaterials and their physiological interactions. Materials used in medicine/dentistry: metals, ceramics, polymers, composites , resorbable smart, natural materials. Material response/degradation: mechanical breakdown, corrosion, dissolution, leaching, chemical degradation, wear. Host responses: foreign body reactions, inflammation, wound healing, carcinogenicity, immunogenicity, cytotoxicty, infection, local/systemic effects.BIOMED 483MRI Magnetic Resonance Imaging BIOMED 483 - MRI Magnetic Resonance Imaging:Introduction to the physics, techniques and applications of magnetic resonance imaging (MRI). Basics of nuclear magnetic resonance physics, spectral analysis and Fourier transforms, techniques for spatial localization, MRI hardware. Applications of MRI including magnetic resonance properties of biological tissues and contrast agents, imaging of anatomy and function.BIOMED 550Ethics & Enterprise
BIOMED 483 - MRI Magnetic Resonance Imaging:Introduction to the physics, techniques and applications of magnetic resonance imaging (MRI). Basics of nuclear magnetic resonance physics, spectral analysis and Fourier transforms, techniques for spatial localization, MRI hardware. Applications of MRI including magnetic resonance properties of biological tissues and contrast agents, imaging of anatomy and function.BIOMED 550Ethics & Enterprise BIOMED 550 - Ethics & Enterprise:Ethics, technology transfer, and technology protection pertaining to Biomedical Engineering are studied . Ethics issues range from the proper research conduct to identifying and managing conflicts of interest. Technology transfer studies the process and its influences on relationships between academia and industry. Technology protection covers legal issues such as patents, copyrights, and contracts.BIOMED 599-105Micro-/Nano-Technology for Biology
BIOMED 550 - Ethics & Enterprise:Ethics, technology transfer, and technology protection pertaining to Biomedical Engineering are studied . Ethics issues range from the proper research conduct to identifying and managing conflicts of interest. Technology transfer studies the process and its influences on relationships between academia and industry. Technology protection covers legal issues such as patents, copyrights, and contracts.BIOMED 599-105Micro-/Nano-Technology for Biology BIOMED 599-105 - Micro-/Nano-Technology for Biology:Course GoalsBIOMED 590Biomedical Engineering ResearchBIOMED 800Biomedical Engineering Graduate Seminar
BIOMED 599-105 - Micro-/Nano-Technology for Biology:Course GoalsBIOMED 590Biomedical Engineering ResearchBIOMED 800Biomedical Engineering Graduate Seminar




 Molecular and Cell Biology [5]ANAT 504Cellular Biotechnology
Molecular and Cell Biology [5]ANAT 504Cellular Biotechnology ANAT 504 - Cellular Biotechnology:The course will combine lectures, literature discussion and student presentations on topics relevant to biotechnology. This class covers a wide range of topics in key scientific areas and methodologies, such as immunotherapy, RNAi, stem cells, systems biology, pharmacokinetics, commercialization strategies and more. During the class, students will also be exposed to concepts relevant to product development and commercialization, as well as scientific topics. Grades are determined by class participation, short take-home assignments, student presentations and a final small-group proposal.ANAT 530Molecular Cell Biology
ANAT 504 - Cellular Biotechnology:The course will combine lectures, literature discussion and student presentations on topics relevant to biotechnology. This class covers a wide range of topics in key scientific areas and methodologies, such as immunotherapy, RNAi, stem cells, systems biology, pharmacokinetics, commercialization strategies and more. During the class, students will also be exposed to concepts relevant to product development and commercialization, as well as scientific topics. Grades are determined by class participation, short take-home assignments, student presentations and a final small-group proposal.ANAT 530Molecular Cell Biology ANAT 530 - Molecular Cell Biology:This graduate course is designed to present basic information as well as the most recent developments in key areas of cell biology. Course consists of both lectures by faculty in their areas of expertise and small discussion groups that delve more deeply into lecture material and discuss primary literature. Both will expose students to current experimental approaches in cell biology. Students will be expected to demonstrate their knowledge of course material by participation in discussion groups and by examinations.CDB 680Organogenesis of Complex Tissues I:Gut
ANAT 530 - Molecular Cell Biology:This graduate course is designed to present basic information as well as the most recent developments in key areas of cell biology. Course consists of both lectures by faculty in their areas of expertise and small discussion groups that delve more deeply into lecture material and discuss primary literature. Both will expose students to current experimental approaches in cell biology. Students will be expected to demonstrate their knowledge of course material by participation in discussion groups and by examinations.CDB 680Organogenesis of Complex Tissues I:Gut CDB 680 - Organogenesis of Complex Tissues I:Gut:The course will cover multiple aspects of organogenesis, including: morphological and molecular events underlying organ formation; quantitative aspects of gradient formation, tissue modeling and cell behavior; in vitro and in vivo experimental systems; parallel pathways for organ formation in various model organisms; adult organ structure and pathology; organ regeneration/repair; stem cell systems: cell and tissue engineering; and carcinogenesis.CDB 681Org. of Complex Tissues II:Neural Crest
CDB 680 - Organogenesis of Complex Tissues I:Gut:The course will cover multiple aspects of organogenesis, including: morphological and molecular events underlying organ formation; quantitative aspects of gradient formation, tissue modeling and cell behavior; in vitro and in vivo experimental systems; parallel pathways for organ formation in various model organisms; adult organ structure and pathology; organ regeneration/repair; stem cell systems: cell and tissue engineering; and carcinogenesis.CDB 681Org. of Complex Tissues II:Neural Crest CDB 680 - Organogenesis of Complex Tissues II:Neural Crest:The course will cover multiple aspects of organogenesis, including: morphological and molecular events underlying organ formation; quantitative aspects of gradient formation, tissue modeling and cell behavior; in vitro and in vivo experimental systems; parallel pathways for organ formation in various model organisms; adult organ structure and pathology; organ regeneration/repair; stem cell systems: cell and tissue engineering; and carcinogenesis.CDB 682Org. of Complex Tissues III:Skeletal Muscle
CDB 680 - Organogenesis of Complex Tissues II:Neural Crest:The course will cover multiple aspects of organogenesis, including: morphological and molecular events underlying organ formation; quantitative aspects of gradient formation, tissue modeling and cell behavior; in vitro and in vivo experimental systems; parallel pathways for organ formation in various model organisms; adult organ structure and pathology; organ regeneration/repair; stem cell systems: cell and tissue engineering; and carcinogenesis.CDB 682Org. of Complex Tissues III:Skeletal Muscle CDB 682 - Org. of Complex Tissues III:Skeletal Muscle:The course will cover multiple aspects of organogenesis, including: morphological and molecular events underlying organ formation; quantitative aspects of gradient formation, tissue modeling and cell behavior; in vitro and in vivo experimental systems; parallel pathways for organ formation in various model organisms; adult organ structure and pathology; organ regeneration/repair; stem cell systems: cell and tissue engineering; and carcinogenesis.Bioinformatics and Genetics [5]HGEN 541Gene Structure and Regulation
CDB 682 - Org. of Complex Tissues III:Skeletal Muscle:The course will cover multiple aspects of organogenesis, including: morphological and molecular events underlying organ formation; quantitative aspects of gradient formation, tissue modeling and cell behavior; in vitro and in vivo experimental systems; parallel pathways for organ formation in various model organisms; adult organ structure and pathology; organ regeneration/repair; stem cell systems: cell and tissue engineering; and carcinogenesis.Bioinformatics and Genetics [5]HGEN 541Gene Structure and Regulation HGEN 541 - Gene Structure and Regulation:A combination of classic and current papers in molecular genetics will be selected to accompany the lecture material (1-2 papers per lecture). The foundations of modern genetics will launch the course, including both the fundamentals and current research methods for analysis of gene structure and gene expression. The gene expression component will include positive and negative regulation of transcription, and mRNA splicing and turnover. The basics of DNA recombination, repair and transposition will be covered in relationship to cancer, evolution and mutagenesis. Strategies for developmental regulation will be presented. Parallels between prokaryotes and eukaryotes will be drawn, and comparisons will be made between the temporal and spatial control of gene expression in vertebrates and invertebrates. Genetic engineering topics will include gene targeting and transgenesis, with applications to understanding tissue specific control of gene expression. The course will close with discussion of the Genome Project, identification of disease genes and an introduction to the medical application of molecular genetics including gene therapy. Prerequisites: Foundations in basic biochemistry and genetics are both required for this course.BIOCHEM 511BioInformatics Seminar
HGEN 541 - Gene Structure and Regulation:A combination of classic and current papers in molecular genetics will be selected to accompany the lecture material (1-2 papers per lecture). The foundations of modern genetics will launch the course, including both the fundamentals and current research methods for analysis of gene structure and gene expression. The gene expression component will include positive and negative regulation of transcription, and mRNA splicing and turnover. The basics of DNA recombination, repair and transposition will be covered in relationship to cancer, evolution and mutagenesis. Strategies for developmental regulation will be presented. Parallels between prokaryotes and eukaryotes will be drawn, and comparisons will be made between the temporal and spatial control of gene expression in vertebrates and invertebrates. Genetic engineering topics will include gene targeting and transgenesis, with applications to understanding tissue specific control of gene expression. The course will close with discussion of the Genome Project, identification of disease genes and an introduction to the medical application of molecular genetics including gene therapy. Prerequisites: Foundations in basic biochemistry and genetics are both required for this course.BIOCHEM 511BioInformatics Seminar BIOCHEM 511 - BioInformatics Seminar:This is a seminar course consisting of attendance at the Parke-Davis/University of Michigan seminar series, "Advances in Genome Sciences", critical reading of related manuscripts prior to the seminar, and participation in roundtable discussions with each of the speakers.BIOCHEM 526Survey in BioInformatics
BIOCHEM 511 - BioInformatics Seminar:This is a seminar course consisting of attendance at the Parke-Davis/University of Michigan seminar series, "Advances in Genome Sciences", critical reading of related manuscripts prior to the seminar, and participation in roundtable discussions with each of the speakers.BIOCHEM 526Survey in BioInformatics BIOCHEM 526 - Survey in BioInformatics:This course consists of a series of integrated lectures providing an introduction to bioinformatics and functional genomics. The course will focus on the basic knowledge required in this field, including the theory and design of databases, access to genome information, sources of data, and tools for data mining. The course will also cover identification of both lower order and higher order informational patterns in DNA and approaches to linking genome data to information on gene function. The participation of nationally recognized scientists having expertise in all aspects of Bioinformatics will be solicited.BIOCHEM 600Lab rotation (genetic regulation lab)
BIOCHEM 526 - Survey in BioInformatics:This course consists of a series of integrated lectures providing an introduction to bioinformatics and functional genomics. The course will focus on the basic knowledge required in this field, including the theory and design of databases, access to genome information, sources of data, and tools for data mining. The course will also cover identification of both lower order and higher order informational patterns in DNA and approaches to linking genome data to information on gene function. The participation of nationally recognized scientists having expertise in all aspects of Bioinformatics will be solicited.BIOCHEM 600Lab rotation (genetic regulation lab) BIOCHEM 600 - Lab rotation (genetic regulation lab):Intensive independent study laboratory course for first-year biochemistry Ph.D. students. Laboratory.BIOCHEM 701Signal Transduction, Regulation, and Development
BIOCHEM 600 - Lab rotation (genetic regulation lab):Intensive independent study laboratory course for first-year biochemistry Ph.D. students. Laboratory.BIOCHEM 701Signal Transduction, Regulation, and Development BIOCHEM 701 - Signal Transduction, Regulation, and Development :Discussion of papers covering recent advances in the control of developmental processes by signal transduction and the regulation of gene expression. Topics from both vertebrate and invertebrate systems will be covered, with the main focus on studies of the developing nervous system. Specific topics and papers vary from year to year.Computer Engineering [4]
BIOCHEM 701 - Signal Transduction, Regulation, and Development :Discussion of papers covering recent advances in the control of developmental processes by signal transduction and the regulation of gene expression. Topics from both vertebrate and invertebrate systems will be covered, with the main focus on studies of the developing nervous system. Specific topics and papers vary from year to year.Computer Engineering [4] Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.Hardware Design and Circuits [10]EECS 216Circuit Analysis
Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.Hardware Design and Circuits [10]EECS 216Circuit Analysis EECS 216 (EECS 211) - Circuit Analysis:Introductory electrical engineering topics, continued:basic circuit analysis; elementary transistor and diode circuits. Equivalent transformations of electric circuits. Transient analysis of circuits. Introduction to diode and transistor circuits. Amplifiers, limiters, filters and logic circuits. Laboratory experience with electrical signals and circuits.EECS 270Logic Design
EECS 216 (EECS 211) - Circuit Analysis:Introductory electrical engineering topics, continued:basic circuit analysis; elementary transistor and diode circuits. Equivalent transformations of electric circuits. Transient analysis of circuits. Introduction to diode and transistor circuits. Amplifiers, limiters, filters and logic circuits. Laboratory experience with electrical signals and circuits.EECS 270Logic Design EECS 270 - Introduction to Logic Design:Binary and non-binary systems, Boolean algebra digital design techniques, logic gates, logic minimization, standard combinational circuits, sequential circuits, flip-flops, synthesis of synchronous sequential circuits, PLA's, ROM's, RAM's, arithmetic circuits, computer-aided design. Laboratory includes hardware design and CAD experiments.EECS 317Transistors and Digital Electronics
EECS 270 - Introduction to Logic Design:Binary and non-binary systems, Boolean algebra digital design techniques, logic gates, logic minimization, standard combinational circuits, sequential circuits, flip-flops, synthesis of synchronous sequential circuits, PLA's, ROM's, RAM's, arithmetic circuits, computer-aided design. Laboratory includes hardware design and CAD experiments.EECS 317Transistors and Digital Electronics EECS 317 (EECS 313) - Solid-State Devices and Electronic:Introduction to semiconductor characteristics and to active devices (diodes, field-effect transistors, and bipolar junction transistors). Large signal circuit analysis and design. Computer-aided design and circuit simulation. Digital logic families. Memory circuits (SRAM, DRAM, and ROM). Lectures, written homework sets and computer-based homework.EECS 370CPU Organization, Architecture & Design
EECS 317 (EECS 313) - Solid-State Devices and Electronic:Introduction to semiconductor characteristics and to active devices (diodes, field-effect transistors, and bipolar junction transistors). Large signal circuit analysis and design. Computer-aided design and circuit simulation. Digital logic families. Memory circuits (SRAM, DRAM, and ROM). Lectures, written homework sets and computer-based homework.EECS 370CPU Organization, Architecture & Design EECS 370 - Introduction to Computer Organization:Computer organization will be presented as a hierarchy of virtual machines representing the different abstractions from which computers can be viewed. These include the logic level, microprogramming level, and assembly language level. Lab experiments will explore the design of a microprogrammed computer.EECS 373Design of MicroProcessor Systems
EECS 370 - Introduction to Computer Organization:Computer organization will be presented as a hierarchy of virtual machines representing the different abstractions from which computers can be viewed. These include the logic level, microprogramming level, and assembly language level. Lab experiments will explore the design of a microprogrammed computer.EECS 373Design of MicroProcessor Systems EECS 373 - Design of Microprocessor-Based Systems:Principles of hardware and software microcomputer interfacing; digital logic design and implementation. Experiments with specially designed laboratory facilities. Introduction to digital development equipment and logic analyzers. Assembly language programming. Lecture and laboratory.EECS 423Solid State Devices Lab
EECS 373 - Design of Microprocessor-Based Systems:Principles of hardware and software microcomputer interfacing; digital logic design and implementation. Experiments with specially designed laboratory facilities. Introduction to digital development equipment and logic analyzers. Assembly language programming. Lecture and laboratory.EECS 423Solid State Devices Lab EECS 423 - Solid State Devices Lab:Semiconductor material and device fabrication and evaluation: diodes, bipolar and field-effect transistors, passive components. Semiconductor processing techniques: oxidation, diffusion, deposition, etching, photolithography. Lecture and laboratory. Projects to design and simulate device fabrication sequence.EECS 427VLSI Design (Very Large Scale Integration)
EECS 423 - Solid State Devices Lab:Semiconductor material and device fabrication and evaluation: diodes, bipolar and field-effect transistors, passive components. Semiconductor processing techniques: oxidation, diffusion, deposition, etching, photolithography. Lecture and laboratory. Projects to design and simulate device fabrication sequence.EECS 427VLSI Design (Very Large Scale Integration) EECS 427 - VLSI Design I:Design techniques for rapid implementations of very large scale integrated (VLSI) circuits, MOS technology and logic. Structured design. Design rules, layout procedures. Design aids:layout, design rule checking, logic and circuit simulation. Timing. Testability. Architectures for VLSI. Projects to develop and lay out circuits.EECS 478Switching Theory and Sequential Systems
EECS 427 - VLSI Design I:Design techniques for rapid implementations of very large scale integrated (VLSI) circuits, MOS technology and logic. Structured design. Design rules, layout procedures. Design aids:layout, design rule checking, logic and circuit simulation. Timing. Testability. Architectures for VLSI. Projects to develop and lay out circuits.EECS 478Switching Theory and Sequential Systems EECS 478 - Logic Circuit Synthesis and Optimization:Advanced design of logic circuits. Technology constraints. Theoretical foundations. Computer-aided design algorithms. Two-level and multilevel optimization of combinational circuits. Optimization of finite-state machines. High-level synthesis techniques:modeling, scheduling, and binding. Verification and testing.EECS 498-1MEMS:Micro Electro Mechanical SystemsEECS 598-1Nano-Electronics and Nano-FabricationComp. Theory and ProgrammingEECS 280C Programming and Data Structures
EECS 478 - Logic Circuit Synthesis and Optimization:Advanced design of logic circuits. Technology constraints. Theoretical foundations. Computer-aided design algorithms. Two-level and multilevel optimization of combinational circuits. Optimization of finite-state machines. High-level synthesis techniques:modeling, scheduling, and binding. Verification and testing.EECS 498-1MEMS:Micro Electro Mechanical SystemsEECS 598-1Nano-Electronics and Nano-FabricationComp. Theory and ProgrammingEECS 280C Programming and Data Structures EECS 280 - Programming and Introductory Data Structures:Techniques and algorithm development and effective programming, top-down analysis, structured programming, testing, and program correctness. Program language syntax and static and run-time semantics. Scope, procedure instantiation, recursion, abstract data types, and parameter passing methods. Structured data types, pointers, linked data structures, stacks, queues, arrays, records, and trees.EECS 303Discrete Structures
EECS 280 - Programming and Introductory Data Structures:Techniques and algorithm development and effective programming, top-down analysis, structured programming, testing, and program correctness. Program language syntax and static and run-time semantics. Scope, procedure instantiation, recursion, abstract data types, and parameter passing methods. Structured data types, pointers, linked data structures, stacks, queues, arrays, records, and trees.EECS 303Discrete Structures EECS 303 - Discrete Structures:Fundamental concepts of algebra; partially ordered sets, lattices, Boolean algebras, semigroups, rings, polynomial rings. Graphical representation of algebraic systems; graphs, directed graphs. Application of these concepts to various areas of computer engineering.EECS 380Data Structures and Algorithms in C++
EECS 303 - Discrete Structures:Fundamental concepts of algebra; partially ordered sets, lattices, Boolean algebras, semigroups, rings, polynomial rings. Graphical representation of algebraic systems; graphs, directed graphs. Application of these concepts to various areas of computer engineering.EECS 380Data Structures and Algorithms in C++ EECS 380 - Data Structures and Algorithms:Abstract data types. Recurrence relations and recursions. Advanced data structures:sparse matrices, generalized lists, strings. Tree-searching algorithms, graph algorithms general searching and sorting. Dynamic storage management. Analysis of algorithms O-notation. Complexity. Top-down program development:design, implementation, testing modularity. Several programming assignments.EECS 400Linear Spaces & Matrix Theory
EECS 380 - Data Structures and Algorithms:Abstract data types. Recurrence relations and recursions. Advanced data structures:sparse matrices, generalized lists, strings. Tree-searching algorithms, graph algorithms general searching and sorting. Dynamic storage management. Analysis of algorithms O-notation. Complexity. Top-down program development:design, implementation, testing modularity. Several programming assignments.EECS 400Linear Spaces & Matrix Theory EECS 400 - Linear Spaces & Matrix Theory:Finite dimensional linear spaces and matrix representations of linear transformations. Bases, subspaces, determinants, eigenvectors, and canonical forms. Structure of solutions of systems of linear equations. Applications to differential and difference equations. The course provides more depth and content than Math. 417. Math. 513 is the proper election for students contemplating research in mathematics.EECS 401Probabilistic Methods in Engineering
EECS 400 - Linear Spaces & Matrix Theory:Finite dimensional linear spaces and matrix representations of linear transformations. Bases, subspaces, determinants, eigenvectors, and canonical forms. Structure of solutions of systems of linear equations. Applications to differential and difference equations. The course provides more depth and content than Math. 417. Math. 513 is the proper election for students contemplating research in mathematics.EECS 401Probabilistic Methods in Engineering EECS 401 - Probabilistic Methods in Engineering:Basic concepts of probability theory. Random variables:discrete, continuous, and conditional probability distributions; averages; independence. Introduction to discrete and continuous random processes:wide sense stationarity, correlation, spectral density.EECS 476Foundations of Theoretical Comp.Sci.
EECS 401 - Probabilistic Methods in Engineering:Basic concepts of probability theory. Random variables:discrete, continuous, and conditional probability distributions; averages; independence. Introduction to discrete and continuous random processes:wide sense stationarity, correlation, spectral density.EECS 476Foundations of Theoretical Comp.Sci. EECS 476 - Foundations of Computer Science:An introduction to computation theory:finite automata, regular languages, pushdown automata, context-free languages, Turing machines, recursive languages and functions, and computational complexity.EECS 574Theorethical Computer Science
EECS 476 - Foundations of Computer Science:An introduction to computation theory:finite automata, regular languages, pushdown automata, context-free languages, Turing machines, recursive languages and functions, and computational complexity.EECS 574Theorethical Computer Science EECS 574 - Theorethical Computer Science:Fundamentals of the theory of computation and complexity theory. Computability, undecidability, and logic. Relations between complexity classes, NP-completeness, P-completeness, and randomized computation. Applications in selected areas such as cryptography, logic programming, theorem proving, approximation of optimization problems, or parallel computing.EECS 587Parallel Programming and Algorithms
EECS 574 - Theorethical Computer Science:Fundamentals of the theory of computation and complexity theory. Computability, undecidability, and logic. Relations between complexity classes, NP-completeness, P-completeness, and randomized computation. Applications in selected areas such as cryptography, logic programming, theorem proving, approximation of optimization problems, or parallel computing.EECS 587Parallel Programming and Algorithms EECS 587 - Parallel Algorithms:The design and analysis of efficient algorithms for parallel computers. Fundamental problem areas, such as sorting, matrix multiplication, and graph theory, are considered for a variety of parallel architectures. Simulations of one architecture by another.EECS 598-4Quantum ComputingEECS 880Software Engineering Technical Seminars
EECS 587 - Parallel Algorithms:The design and analysis of efficient algorithms for parallel computers. Fundamental problem areas, such as sorting, matrix multiplication, and graph theory, are considered for a variety of parallel architectures. Simulations of one architecture by another.EECS 598-4Quantum ComputingEECS 880Software Engineering Technical Seminars




 Computer Applications [8]EECS 482Operating Systems
Computer Applications [8]EECS 482Operating Systems EECS 482 - Operating Systems:Operating system functions and implementations:multi-tasking; concurrency and synchronization; deadlock; scheduling; resource allocation; real and virtual memory management; input/output; file systems. Students write several substantial programs dealing with concurrency and synchronization in a multitask environment.EECS 489Computer Networks
EECS 482 - Operating Systems:Operating system functions and implementations:multi-tasking; concurrency and synchronization; deadlock; scheduling; resource allocation; real and virtual memory management; input/output; file systems. Students write several substantial programs dealing with concurrency and synchronization in a multitask environment.EECS 489Computer Networks EECS 489 - Computer Networks:Protocols and architectures of computer networks. Topics include client-server computing, socket programming, naming and addressing, media access protocols, routing and transport protocols, flow and congestion control, and other application-specific protocols. Emphasis is placed on understanding protocol design principles. Programming problems to explore design choices and actual implementation issues assigned.EECS 498-2Virtual RealityEECS 498-3Web and InternetEECS 547Electronic Commerce
EECS 489 - Computer Networks:Protocols and architectures of computer networks. Topics include client-server computing, socket programming, naming and addressing, media access protocols, routing and transport protocols, flow and congestion control, and other application-specific protocols. Emphasis is placed on understanding protocol design principles. Programming problems to explore design choices and actual implementation issues assigned.EECS 498-2Virtual RealityEECS 498-3Web and InternetEECS 547Electronic Commerce EECS 547 - Electronic Commerce:The Internet is rapidly changing the way we trade with one another, conduct businesses, and organize financial institutions. In this course, we will cover a range of important principles -- drawn from computer science, economics, and other disciplines -- that influence the design and analysis of Internet commerce systems. We will begin with an introduction to design and analysis methods to make e-Commerce systems robust against failures, malicious attackers, and strategic manipulation. We will then study three aspects of electronic commerce systems: locating buyers and sellers (search), setting terms of trade (negotiation), and verifying and consummating the deal (exchange). The goal is to develop a mastery of the fundamental concepts and approaches through examples, rather than an exhaustive survey of the field. The course is a semester-long, 3-credit course.EECS 582Advanced Operating Systems
EECS 547 - Electronic Commerce:The Internet is rapidly changing the way we trade with one another, conduct businesses, and organize financial institutions. In this course, we will cover a range of important principles -- drawn from computer science, economics, and other disciplines -- that influence the design and analysis of Internet commerce systems. We will begin with an introduction to design and analysis methods to make e-Commerce systems robust against failures, malicious attackers, and strategic manipulation. We will then study three aspects of electronic commerce systems: locating buyers and sellers (search), setting terms of trade (negotiation), and verifying and consummating the deal (exchange). The goal is to develop a mastery of the fundamental concepts and approaches through examples, rather than an exhaustive survey of the field. The course is a semester-long, 3-credit course.EECS 582Advanced Operating Systems EECS 582 - Advanced Operating Systems:The course catalog states, "Course discusses advanced topics and research issues in operating systems. Topics will be drawn from a variety of operating systems areas such as distributed systems and languages, networking, security, and protection, real-time systems, modeling and analysis, etc."
EECS 582 - Advanced Operating Systems:The course catalog states, "Course discusses advanced topics and research issues in operating systems. Topics will be drawn from a variety of operating systems areas such as distributed systems and languages, networking, security, and protection, real-time systems, modeling and analysis, etc."
This semester, we will take a slightly broader view of systems research and consider many common issues that emerge across operating systems, database systems, networked systems, distributed systems, mobile systems, and embedded systems.
The design of computer systems -- whether building-size or hand-held, file system or database -- faces many common challenges and pitfalls. Fortunately, many of the principles and practices are often common as well. This class will focus on identifying and understanding the enduring principles and practices in computer systems design and implementation, and will prepare students to carry our substantial independent systems research projects.EECS 584Advanced Databases EECS584 - Advanced Databases:Survey of advanced topics in database systems. Distributed databases, query processing, transaction provessing. Effects of data models:object-oriented and deductive databases; architectures:main-memory and parallel repositories; distributed organizations:client-server and heterogeneous systems. Basic data management for emerging areas:internet applications, OLAP, data mining. Case studies of existing systems. Group projects.EECS 598-2CryptographyMachine Learning and A.I. [8]EECS 492Artificial Intelligence
EECS584 - Advanced Databases:Survey of advanced topics in database systems. Distributed databases, query processing, transaction provessing. Effects of data models:object-oriented and deductive databases; architectures:main-memory and parallel repositories; distributed organizations:client-server and heterogeneous systems. Basic data management for emerging areas:internet applications, OLAP, data mining. Case studies of existing systems. Group projects.EECS 598-2CryptographyMachine Learning and A.I. [8]EECS 492Artificial Intelligence EECS 492 - Introduction to Artificial Intelligence:Fundamental concepts of AI, organized around the task of building computational agents. Core topics include search, logic, representation and reasoning, automated planning, decision making under uncertainty, and machine learning.EECS 499Genetic ProgrammingEECS 543Knowledge Systems
EECS 492 - Introduction to Artificial Intelligence:Fundamental concepts of AI, organized around the task of building computational agents. Core topics include search, logic, representation and reasoning, automated planning, decision making under uncertainty, and machine learning.EECS 499Genetic ProgrammingEECS 543Knowledge Systems EECS 543 - Knowledge-Based Systems:Techniques and principles for developing application software based on explicit representation and manipulation of domain knowledge, as applied to computer vision, robotic control, design and manufacturing, diagnostics, autonomous systems, etc. Topics include:identifying and representing knowledge, integrating knowledge-based behavior into complex systems, reasoning, and handling uncertainty and unpredictability.EECS 592Advanced Artificial Intelligence
EECS 543 - Knowledge-Based Systems:Techniques and principles for developing application software based on explicit representation and manipulation of domain knowledge, as applied to computer vision, robotic control, design and manufacturing, diagnostics, autonomous systems, etc. Topics include:identifying and representing knowledge, integrating knowledge-based behavior into complex systems, reasoning, and handling uncertainty and unpredictability.EECS 592Advanced Artificial Intelligence EECS 592 - Advanced Artificial Intelligence:Advanced topics in artificial intelligence. Issues in knowledge representation, knowledge based systems, problem solving, planning and other topics will be discussed. Students will work on several projects.EECS 594Adaptive Systems
EECS 592 - Advanced Artificial Intelligence:Advanced topics in artificial intelligence. Issues in knowledge representation, knowledge based systems, problem solving, planning and other topics will be discussed. Students will work on several projects.EECS 594Adaptive Systems EECS 594 - Introduction to Adaptive Systems:Programs and automata that "learn" by adapting to their environment; programs that utilize genetic algorithms for learning. Samuel strategies, realistic neural networks, connectionist systems, classifier systems, and related models of cognition. Artificial intelligence systems, such as NETL and SOAR, are examined for their impact upon machine learning and cognitive science.EECS 599Intelligent Agents Plan RecognitionEECS 695Neural Models
EECS 594 - Introduction to Adaptive Systems:Programs and automata that "learn" by adapting to their environment; programs that utilize genetic algorithms for learning. Samuel strategies, realistic neural networks, connectionist systems, classifier systems, and related models of cognition. Artificial intelligence systems, such as NETL and SOAR, are examined for their impact upon machine learning and cognitive science.EECS 599Intelligent Agents Plan RecognitionEECS 695Neural Models EECS 695 - Neural Models: Mechanisms of Learning:Learning is not only what education is about but also a basic activity of enormous importance to our species. Unfortunately even many whose work depends upon it do not fully grasp the way the mind and environment interact in acquiring, understanding, and storing information. The course focuses on these themes both at a practical level and by exploring the behind-the-scenes activities of neurons that make learning possible. Four major challenges will be studied:PSYC 808Cognitive Modeling
EECS 695 - Neural Models: Mechanisms of Learning:Learning is not only what education is about but also a basic activity of enormous importance to our species. Unfortunately even many whose work depends upon it do not fully grasp the way the mind and environment interact in acquiring, understanding, and storing information. The course focuses on these themes both at a practical level and by exploring the behind-the-scenes activities of neurons that make learning possible. Four major challenges will be studied:PSYC 808Cognitive Modeling PSYCH 808 - Cognitive Modeling:Computing in Cognitive Science.
PSYCH 808 - Cognitive Modeling:Computing in Cognitive Science.
ARCHITECTURES/APPROACHES:Other Fields Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.Exercise Physiology and Kinesiology [6]KIN 503Legal Aspects of Sport
Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.Exercise Physiology and Kinesiology [6]KIN 503Legal Aspects of Sport KIN 503 - Legal Aspects of Sport:This is a comprehensive review of legal aspects affecting sport, recreation, and fitness industries. The range of review includes civil procedure; contracts: employment, leases, waivers; tort liability for coaches, administrators, employees, and independent contractors; 14th Amendment Due Process and Equal Protection; product liability; and statutory regulation including Title VII, Title IX, ADA, Anti-Trust, and IRS codeKIN 520Seminar in Motor Control
KIN 503 - Legal Aspects of Sport:This is a comprehensive review of legal aspects affecting sport, recreation, and fitness industries. The range of review includes civil procedure; contracts: employment, leases, waivers; tort liability for coaches, administrators, employees, and independent contractors; 14th Amendment Due Process and Equal Protection; product liability; and statutory regulation including Title VII, Title IX, ADA, Anti-Trust, and IRS codeKIN 520Seminar in Motor Control KIN 520 - Seminar in Motor Control:Focuses on current issues in movement control from either a neurophysiological or behavioral viewpoint. Students will present assigned readings and will write a paper on an approved topicMVS 542Exercise Physiology and Nutrition
KIN 520 - Seminar in Motor Control:Focuses on current issues in movement control from either a neurophysiological or behavioral viewpoint. Students will present assigned readings and will write a paper on an approved topicMVS 542Exercise Physiology and Nutrition MVS 542 - Exercise Physiology and Nutrition:Biochemical and physiological processes of fuel mobilization and storage in response to exercise and the modification of those processes by nutritional variablesKIN 610Current Issues in KinesiologyKIN 615Phil. of Science and Res. in Kines.
MVS 542 - Exercise Physiology and Nutrition:Biochemical and physiological processes of fuel mobilization and storage in response to exercise and the modification of those processes by nutritional variablesKIN 610Current Issues in KinesiologyKIN 615Phil. of Science and Res. in Kines. KIN 615 - Phil. of Science and Res. in Kines.:Topics include the nature of scientific inquiry, theories of knowledge acquisition; empirical vs. theoretical research; basic vs. applied research; induction and deduction; doubts and alternatives; objectivity of science; facts, laws and theories; pseudo-science; causation and mechanism; formulation of problems, research design and use of statisticsKIN 682Muscle Growth, Strength & Training StrategiesAerospace Engineering [5]AERO 453Prob. Methods in Engineering
KIN 615 - Phil. of Science and Res. in Kines.:Topics include the nature of scientific inquiry, theories of knowledge acquisition; empirical vs. theoretical research; basic vs. applied research; induction and deduction; doubts and alternatives; objectivity of science; facts, laws and theories; pseudo-science; causation and mechanism; formulation of problems, research design and use of statisticsKIN 682Muscle Growth, Strength & Training StrategiesAerospace Engineering [5]AERO 453Prob. Methods in Engineering AERO 453 - Probabilistic Methods in Engineering:Basic concepts of probability theory. Random variables: discrete, continuous, and conditional probability distributions; averages; independence. Statistical inference: hypothesis testing and estimation. Introduction to discrete and continuous random processes.TECH-COMM 498Technical Writing & Comm.
AERO 453 - Probabilistic Methods in Engineering:Basic concepts of probability theory. Random variables: discrete, continuous, and conditional probability distributions; averages; independence. Statistical inference: hypothesis testing and estimation. Introduction to discrete and continuous random processes.TECH-COMM 498Technical Writing & Comm. TECH-COMM 498 - Technical Writing and Communication:Technical and Professional Writing for Industry, Government, and Business. Senior or graduate standing.AERO 582Spacecraft Technology
TECH-COMM 498 - Technical Writing and Communication:Technical and Professional Writing for Industry, Government, and Business. Senior or graduate standing.AERO 582Spacecraft Technology AERO 582 - Spacecraft Technology:Systematic and comprehensive review of spacecraft and space mission design and key technologies for space missions. Discussions on project management and the economic and political factors that affect space missions. Specific space mission designs are developed in teams.AERO 590Aerospace Research
AERO 582 - Spacecraft Technology:Systematic and comprehensive review of spacecraft and space mission design and key technologies for space missions. Discussions on project management and the economic and political factors that affect space missions. Specific space mission designs are developed in teams.AERO 590Aerospace Research AERO 590 - Aerospace Research:Study of advanced aspects of aerospace engineering directed by an Aerospace faculty member. Primarily for graduates. The student will submit a final report.AERO 597Space Plasma Physics
AERO 590 - Aerospace Research:Study of advanced aspects of aerospace engineering directed by an Aerospace faculty member. Primarily for graduates. The student will submit a final report.AERO 597Space Plasma Physics AERO 597 - Space Plasma Physics:Basic plasma concepts, Boltzmann equation, higher order moments equations, MHD equations, double adiabatic theory. Plasma expansion to vacuum, transonic flows, solar wind, polar wind. Collisionless shocks, propagating and planetary shocks. Fokker-Planck equation, quasilinear theory, velocity diffusion, cosmic ray transport, shock acceleration. Spacecraft charging, mass loading.EconomicsECON 501Micro-Economic Theory
AERO 597 - Space Plasma Physics:Basic plasma concepts, Boltzmann equation, higher order moments equations, MHD equations, double adiabatic theory. Plasma expansion to vacuum, transonic flows, solar wind, polar wind. Collisionless shocks, propagating and planetary shocks. Fokker-Planck equation, quasilinear theory, velocity diffusion, cosmic ray transport, shock acceleration. Spacecraft charging, mass loading.EconomicsECON 501Micro-Economic Theory ECON 501 - Applied Microeconomic Theory:A course designed for students in the MAE program. Basic models in the principal areas of microeconomic theory are covered: consumer demand, production and costs, product markets, factor markets, allocative efficiency, and corrections for market failure. Most of the course is spent studying the use of these tools in the analysis of specific microeconomic policy problems. Application of theory to current policy problems is stressed, and a substantial amount of class time is devoted to exercises based on such problems.
ECON 501 - Applied Microeconomic Theory:A course designed for students in the MAE program. Basic models in the principal areas of microeconomic theory are covered: consumer demand, production and costs, product markets, factor markets, allocative efficiency, and corrections for market failure. Most of the course is spent studying the use of these tools in the analysis of specific microeconomic policy problems. Application of theory to current policy problems is stressed, and a substantial amount of class time is devoted to exercises based on such problems.





Science and Philosophy [5] Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.Math [9]MATH 115Calculus I
Course Title:No course selected. Please move the cursor over a course title to select one.Course Description:Description of the course content will be displayed here instantaneously by hovering above any course title.Math [9]MATH 115Calculus I MATH 115 - Calculus I:The sequence Math 115-116-215 is the standard complete introduction to the concepts and methods of calculus. It is taken by the majority of students intending to concentrate in mathematics, science, or engineering as well as students heading for many other fields. The emphasis is on concepts and solving problems rather than theory and proof. All sections are given a uniform midterm and final exam. The course presents the concepts of calculus from three points of view: geometric (graphs); numerical (tables); and algebraic (formulas). Students will develop their reading, writing, and questioning skills.MATH 116Calculus II
MATH 115 - Calculus I:The sequence Math 115-116-215 is the standard complete introduction to the concepts and methods of calculus. It is taken by the majority of students intending to concentrate in mathematics, science, or engineering as well as students heading for many other fields. The emphasis is on concepts and solving problems rather than theory and proof. All sections are given a uniform midterm and final exam. The course presents the concepts of calculus from three points of view: geometric (graphs); numerical (tables); and algebraic (formulas). Students will develop their reading, writing, and questioning skills.MATH 116Calculus II MATH 116 - Calculus II:Topics include the indefinite integral, techniques of integration, introduction to differential equations, infinite series.MATH 215Calculus III
MATH 116 - Calculus II:Topics include the indefinite integral, techniques of integration, introduction to differential equations, infinite series.MATH 215Calculus III MATH 215 - Calculus III:Topics include vector algebra and vector functions; analytic geometry of planes, surfaces, and solids; functions of several variables and partial differentiation; line, surface, and volume integrals and applications; vector fields and integration; Green's Theorem and Stokes' Theorem.MATH 216Differential Eq.
MATH 215 - Calculus III:Topics include vector algebra and vector functions; analytic geometry of planes, surfaces, and solids; functions of several variables and partial differentiation; line, surface, and volume integrals and applications; vector fields and integration; Green's Theorem and Stokes' Theorem.MATH 216Differential Eq. MATH 216 - Differential Equations:After an introduction to ordinary differential equations, the first half of the course is devoted to topics in linear algebra, including systems of linear algebraic equations, vector spaces, linear dependence, bases, dimension, matrix algebra, determinants, eigenvalues, and eigenvectors. In the second half these tools are applied to the solution of linear systems of ordinary differential equations. Topics include: oscillating systems, the Laplace transform, initial value problems, resonance, phase portraits, and an introduction to numerical methods.STAT 402Statistics and Data Analysis
MATH 216 - Differential Equations:After an introduction to ordinary differential equations, the first half of the course is devoted to topics in linear algebra, including systems of linear algebraic equations, vector spaces, linear dependence, bases, dimension, matrix algebra, determinants, eigenvalues, and eigenvectors. In the second half these tools are applied to the solution of linear systems of ordinary differential equations. Topics include: oscillating systems, the Laplace transform, initial value problems, resonance, phase portraits, and an introduction to numerical methods.STAT 402Statistics and Data Analysis STAT 402 - Statistics and Data Analysis:In this course students are introduced to the concepts and applications of statistical methods and data analysis. Examples of applications are drawn from virtually all academic areas and some attention is given to statistical process control methods. The course format includes lectures (3 hours per week) and a laboratory (l.5 hours per week). The laboratory section deals with the computational aspects of the course and provides a forum for review of lecture material. For this purpose, students are introduced to the use of a statistical analysis-computer package.MATH 419Honors Vector Spaces and Matrix Theory
STAT 402 - Statistics and Data Analysis:In this course students are introduced to the concepts and applications of statistical methods and data analysis. Examples of applications are drawn from virtually all academic areas and some attention is given to statistical process control methods. The course format includes lectures (3 hours per week) and a laboratory (l.5 hours per week). The laboratory section deals with the computational aspects of the course and provides a forum for review of lecture material. For this purpose, students are introduced to the use of a statistical analysis-computer package.MATH 419Honors Vector Spaces and Matrix Theory MATH 419 - Honors Vector Spaces and Matrix Theory:Linear equations, Gauss-Jordan elimination, linear transformations and their inverses, matrix algebra, subspaces, linear independence, bases, orthogonality, Gram-Schmidt, orthogonal transformations and matrices, least squares, determinants, eigenvalues and eigenvectors, coordinate systems, diagonalization, and quadratic forms.MATH 425Probability
MATH 419 - Honors Vector Spaces and Matrix Theory:Linear equations, Gauss-Jordan elimination, linear transformations and their inverses, matrix algebra, subspaces, linear independence, bases, orthogonality, Gram-Schmidt, orthogonal transformations and matrices, least squares, determinants, eigenvalues and eigenvectors, coordinate systems, diagonalization, and quadratic forms.MATH 425Probability MATH 425 - Introduction to Probability:This course introduces students to useful and interesting ideas of the mathematical theory of probability and to a number of applications of probability to a variety of fields including genetics, economics, geology, business, and engineering. The theory developed together with other mathematical tools such as combinatorics and calculus are applied to everyday problems. Concepts, calculations, and derivations are emphasized. The course will make essential use of the material of Math 116 and 215. Math concentrators should be sure to elect sections of the course that are taught by Mathematics (not Statistics) faculty. Topics include the basic results and methods of both discrete and continuous probability theory:conditional probability, independent events, random variables, jointly distributed random variables, expectations, variances, covariances.AERO 453Prob. Methods in Engineering
MATH 425 - Introduction to Probability:This course introduces students to useful and interesting ideas of the mathematical theory of probability and to a number of applications of probability to a variety of fields including genetics, economics, geology, business, and engineering. The theory developed together with other mathematical tools such as combinatorics and calculus are applied to everyday problems. Concepts, calculations, and derivations are emphasized. The course will make essential use of the material of Math 116 and 215. Math concentrators should be sure to elect sections of the course that are taught by Mathematics (not Statistics) faculty. Topics include the basic results and methods of both discrete and continuous probability theory:conditional probability, independent events, random variables, jointly distributed random variables, expectations, variances, covariances.AERO 453Prob. Methods in Engineering AERO 453 - Prob. Methods in Engineering:Basic concepts of probability theory. Random variables: discrete, continuous, and conditional probability distributions; averages; independence. Statistical inference: hypothesis testing and estimation. Introduction to discrete and continuous random processes.MATH 550Intro Adaptive Systems
AERO 453 - Prob. Methods in Engineering:Basic concepts of probability theory. Random variables: discrete, continuous, and conditional probability distributions; averages; independence. Statistical inference: hypothesis testing and estimation. Introduction to discrete and continuous random processes.MATH 550Intro Adaptive Systems MATH 550 - Intro Adaptive Systems:Intro to Adaptive Systems (aka "Introduction to Evolutionary Dynamical Systems") centers on the construction and use of agent-based adaptive models study phenomena which are prototypical in the social, biological and decision sciences. These models are "agent-based" or "bottom-up" in that the structure placed at the le vel of the individuals as basic components; they are "adaptive" in that individuals often adapt to their environment through evolution or learning. The goal of these models is to understand how the structure at the individual or micro level leads to emerg ent behavior at the macro or aggregate level. Often the individuals are grouped into subpopulations or interesting hierarchies, and the researcher may want to understand how the structure of development of these populations affects macroscopic outcomes. The course will start with classical differential equation and game theory approaches. It will then focus on the theory and application of particular models of adaptive systems such as models of neural systems, genetic algorithms, classifier system and cel lular automata. Time permitting, we will discuss more recent developments such as sugarscape and echo.Philosophy [9]PHIL 1x1General Philosophy IPHIL 2x1General Philosophy IIPHIL 356Issues in Bio-Ethics
MATH 550 - Intro Adaptive Systems:Intro to Adaptive Systems (aka "Introduction to Evolutionary Dynamical Systems") centers on the construction and use of agent-based adaptive models study phenomena which are prototypical in the social, biological and decision sciences. These models are "agent-based" or "bottom-up" in that the structure placed at the le vel of the individuals as basic components; they are "adaptive" in that individuals often adapt to their environment through evolution or learning. The goal of these models is to understand how the structure at the individual or micro level leads to emerg ent behavior at the macro or aggregate level. Often the individuals are grouped into subpopulations or interesting hierarchies, and the researcher may want to understand how the structure of development of these populations affects macroscopic outcomes. The course will start with classical differential equation and game theory approaches. It will then focus on the theory and application of particular models of adaptive systems such as models of neural systems, genetic algorithms, classifier system and cel lular automata. Time permitting, we will discuss more recent developments such as sugarscape and echo.Philosophy [9]PHIL 1x1General Philosophy IPHIL 2x1General Philosophy IIPHIL 356Issues in Bio-Ethics PHIL 356 - Issues in Bio-Ethics:This course will focus on moral problems that arise in biomedical ethics. Biomedical ethics is composed of two separate fields: bioethics and medical ethics. Bioethics is the study of the ethics of life (and death), and includes familiar topics such as abortion, cloning, stem cell research, allocation of scarce medical resources, and euthanasia. We shall spend approximately the first two-third of the course on these issues. For the last third of the course, we shall discuss topics in medical ethics, which is concerned with "micro" issues such as the moral underpinnings of doctor-patient relationships as well as "macro" issues such as the structures of medical institutions or the duties that societies have to provide health care for those in need. No previous coursework in philosophy is required for this course and fundamental concepts in moral philosophy (e.g., consequentialism and deontology) will be explained as they become relevant. This is a course on theoretical (as opposed to clinical) bioethics.PHIL 423Space, Time & Einstein's Relativity
PHIL 356 - Issues in Bio-Ethics:This course will focus on moral problems that arise in biomedical ethics. Biomedical ethics is composed of two separate fields: bioethics and medical ethics. Bioethics is the study of the ethics of life (and death), and includes familiar topics such as abortion, cloning, stem cell research, allocation of scarce medical resources, and euthanasia. We shall spend approximately the first two-third of the course on these issues. For the last third of the course, we shall discuss topics in medical ethics, which is concerned with "micro" issues such as the moral underpinnings of doctor-patient relationships as well as "macro" issues such as the structures of medical institutions or the duties that societies have to provide health care for those in need. No previous coursework in philosophy is required for this course and fundamental concepts in moral philosophy (e.g., consequentialism and deontology) will be explained as they become relevant. This is a course on theoretical (as opposed to clinical) bioethics.PHIL 423Space, Time & Einstein's Relativity PHIL 423 - Problems of Space and Time:Traditional philosophical questions about the nature of time and space have been strikingly influenced in the twentieth century by the results of contemporary physical science. At the same time, the important current physical theories of space and time rest explicitly or implicitly on deep-rooted philosophical assumptions. The purpose of this course is to study the mutual interaction between science and philosophy as illustrated in problems about space and time. Typical topics to be considered include the status of knowledge about the structure of space and time, substantial versus relational theories of space-time, spatio-temporal order and causal order, and the so-called problem of the direction of time. This course can be appreciated by students who have either a background in philosophy - especially logic and philosophy of science, metaphysics, epistemology - or background in physical science or mathematics. An attempt is made in this course to introduce the fundamental ideas of both philosophy and science at a level which can be understood by those without extensive background so students need not be proficient in both science and philosophy to benefit from the course. The primary text is L. Sklar Space, Time, and Spacetime. There are additional readings from such authors as Reichenbach, Poincaré, Grunbaum, Smart, Wheeler, and others.PHIL 429Ethical Analysis
PHIL 423 - Problems of Space and Time:Traditional philosophical questions about the nature of time and space have been strikingly influenced in the twentieth century by the results of contemporary physical science. At the same time, the important current physical theories of space and time rest explicitly or implicitly on deep-rooted philosophical assumptions. The purpose of this course is to study the mutual interaction between science and philosophy as illustrated in problems about space and time. Typical topics to be considered include the status of knowledge about the structure of space and time, substantial versus relational theories of space-time, spatio-temporal order and causal order, and the so-called problem of the direction of time. This course can be appreciated by students who have either a background in philosophy - especially logic and philosophy of science, metaphysics, epistemology - or background in physical science or mathematics. An attempt is made in this course to introduce the fundamental ideas of both philosophy and science at a level which can be understood by those without extensive background so students need not be proficient in both science and philosophy to benefit from the course. The primary text is L. Sklar Space, Time, and Spacetime. There are additional readings from such authors as Reichenbach, Poincaré, Grunbaum, Smart, Wheeler, and others.PHIL 429Ethical Analysis PHIL 429 - Ethical Analysis:Questions about the nature and standing of morality arise in both theory and practice. Moreover, in recent years morality has served as a central example in wide philosophical debates about the nature of normativity and the relation of theory to practice. In this course we will critically investigate several of the most influential philosophical conceptions of morality, including historical as well as contemporary writings. Among the questions we will consider:PHIL 433History of Ethics
PHIL 429 - Ethical Analysis:Questions about the nature and standing of morality arise in both theory and practice. Moreover, in recent years morality has served as a central example in wide philosophical debates about the nature of normativity and the relation of theory to practice. In this course we will critically investigate several of the most influential philosophical conceptions of morality, including historical as well as contemporary writings. Among the questions we will consider:PHIL 433History of Ethics PHIL 433 - History of Ethics:The modern period in moral philosophy began with Thomas Hobbes, whose Leviathan (1651) shook the traditional foundations of ethics and forced those who would defend ethics against (what they saw to be) Hobbes' nihilism to do so in a broadly naturalistic framework that took serious account of recent advances in science. Thus began a period of exciting and fruitful moral philosophy that stretched through the end of the eighteenth century and into the nineteenth. Indeed, even debates now current in moral philosophy almost always can be traced back to origins in this period. This course will be a study of several of the central writers and texts of this "enlightenment" and post-enlightenment period. In addition to Hobbes, we shall read some of Hutcheson, Butler, Hume, Kant, Bentham, Rousseau, Nietzsche, Fichte, and Hegel. We shall end with a radical critic of this broad tradition: Nietzsche. Course requirements: short paper, long paper, final exam.PHIL 602Philosophy of Science
PHIL 433 - History of Ethics:The modern period in moral philosophy began with Thomas Hobbes, whose Leviathan (1651) shook the traditional foundations of ethics and forced those who would defend ethics against (what they saw to be) Hobbes' nihilism to do so in a broadly naturalistic framework that took serious account of recent advances in science. Thus began a period of exciting and fruitful moral philosophy that stretched through the end of the eighteenth century and into the nineteenth. Indeed, even debates now current in moral philosophy almost always can be traced back to origins in this period. This course will be a study of several of the central writers and texts of this "enlightenment" and post-enlightenment period. In addition to Hobbes, we shall read some of Hutcheson, Butler, Hume, Kant, Bentham, Rousseau, Nietzsche, Fichte, and Hegel. We shall end with a radical critic of this broad tradition: Nietzsche. Course requirements: short paper, long paper, final exam.PHIL 602Philosophy of Science PHIL 602 - Philosophy of Science:Our fundamental physical theories are models of mathematical rigor and precision. But how do they actually function in our attempts to give descriptions and explanations of our complex and messy world? Are the lawlike assertions of these theories 'true, ' or falsehoods that carry with them clauses restricting their applicability that can never be explicitly filled out? Or are the laws intended to be true not of the world but only of idealized 'models' of it? Are the basic laws applicable to all physical systems, or only to very limited domains of specially prepared systems in the laboratory? Are we supposed to believe the theories, or, rather, only to believe them to be false but somehow useful stepping stones in an ongoing process in which each successive theory is ultimately rejected as a transient falsehood? Can we understand our theories at face value, or must theories always be accompanied by 'interpretations' that are outside the theory but essential to grasp its import? Are these interpretations themselves part of the scientific process or are they imposed upon the theories somehow from the 'outside?' Do the answers to all of these questions force us to an 'instrumentalist' as opposed to 'realist' understanding of our basic theories?PHIL 611Current Philosophy - The SelfPHIL 615Philosophy of Language
PHIL 602 - Philosophy of Science:Our fundamental physical theories are models of mathematical rigor and precision. But how do they actually function in our attempts to give descriptions and explanations of our complex and messy world? Are the lawlike assertions of these theories 'true, ' or falsehoods that carry with them clauses restricting their applicability that can never be explicitly filled out? Or are the laws intended to be true not of the world but only of idealized 'models' of it? Are the basic laws applicable to all physical systems, or only to very limited domains of specially prepared systems in the laboratory? Are we supposed to believe the theories, or, rather, only to believe them to be false but somehow useful stepping stones in an ongoing process in which each successive theory is ultimately rejected as a transient falsehood? Can we understand our theories at face value, or must theories always be accompanied by 'interpretations' that are outside the theory but essential to grasp its import? Are these interpretations themselves part of the scientific process or are they imposed upon the theories somehow from the 'outside?' Do the answers to all of these questions force us to an 'instrumentalist' as opposed to 'realist' understanding of our basic theories?PHIL 611Current Philosophy - The SelfPHIL 615Philosophy of Language




 Chemistry and BioChemistry [11]CHEM 125General Chemistry and ReactivityCHEM 126General Chemistry and Reactivity LabCHEM 130General and Inorganic Chemistry I & Lab
Chemistry and BioChemistry [11]CHEM 125General Chemistry and ReactivityCHEM 126General Chemistry and Reactivity LabCHEM 130General and Inorganic Chemistry I & Lab CHEM 130 - General and Inorganic Chemistry I:Chemistry 130 provides an introduction to the major concepts of chemistry, including the microscopic picture of atomic and molecular structure, periodic trends in the chemical reactivity, the energetics of chemical reactions, and the nature of chemical equilibria. Students will be introduced to the fundamental principles of modern chemistry, the descriptive chemistry of the elements, and to the underlying theories that account for observed macroscopic behavior. In Chem 130, students will learn to think critically, examine experimental data, and form generalizations about data as chemists do.CHEM 210Organic Chemistry
CHEM 130 - General and Inorganic Chemistry I:Chemistry 130 provides an introduction to the major concepts of chemistry, including the microscopic picture of atomic and molecular structure, periodic trends in the chemical reactivity, the energetics of chemical reactions, and the nature of chemical equilibria. Students will be introduced to the fundamental principles of modern chemistry, the descriptive chemistry of the elements, and to the underlying theories that account for observed macroscopic behavior. In Chem 130, students will learn to think critically, examine experimental data, and form generalizations about data as chemists do.CHEM 210Organic Chemistry CHEM 210 - Organic Chemistry:Chemistry 210 is the first course in a two-term sequence in which the major concepts of chemistry are introduced in the context of organic chemistry. Emphasis is on the development of the capacity of students to think about the relationship between structure and reactivity and to solve problems in a qualitatively analytical way. This course is a particularly good first course for students with AP credit in chemistry, Honors students, and other students with a strong interest in chemistry and biology.CHEM 211Organic Chemistry Lab
CHEM 210 - Organic Chemistry:Chemistry 210 is the first course in a two-term sequence in which the major concepts of chemistry are introduced in the context of organic chemistry. Emphasis is on the development of the capacity of students to think about the relationship between structure and reactivity and to solve problems in a qualitatively analytical way. This course is a particularly good first course for students with AP credit in chemistry, Honors students, and other students with a strong interest in chemistry and biology.CHEM 211Organic Chemistry Lab CHEM 210 - Organic Chemistry Lab:Chemistry 211 is a laboratory introduction to methods of investigation in inorganic and organic chemistry. Students solve individual problems using microscale equipment and a variety of techniques such as thin layer chromatography, titrations, and spectroscopy. The course consists of a four-hour laboratory period with a teaching assistant under the supervision of the professor.BIOCHEM 451Intro Biochemistry
CHEM 210 - Organic Chemistry Lab:Chemistry 211 is a laboratory introduction to methods of investigation in inorganic and organic chemistry. Students solve individual problems using microscale equipment and a variety of techniques such as thin layer chromatography, titrations, and spectroscopy. The course consists of a four-hour laboratory period with a teaching assistant under the supervision of the professor.BIOCHEM 451Intro Biochemistry BIOCHEM 451 - Intro Biochemistry:A rigorous introduction to biochemistry with a chemical emphasis. Designed for undergraduates in the Biochemistry Concentration Program but open to graduate students with a strong background in chemistry. Prerequisites: Chem 215, Biol. 152 or 195, and Math 115.BIOCHEM 516Biochemistry Lab
BIOCHEM 451 - Intro Biochemistry:A rigorous introduction to biochemistry with a chemical emphasis. Designed for undergraduates in the Biochemistry Concentration Program but open to graduate students with a strong background in chemistry. Prerequisites: Chem 215, Biol. 152 or 195, and Math 115.BIOCHEM 516Biochemistry Lab N/A:...BIOCHEM 570Protein Structure
N/A:...BIOCHEM 570Protein Structure N/A:...BIOCHEM 571DNA & Nucleic Acids
N/A:...BIOCHEM 571DNA & Nucleic Acids N/A:...BIOCHEM 572Gene Expression
N/A:...BIOCHEM 572Gene Expression N/A:...BIOCHEM 573Enzyme Kinetics
N/A:...BIOCHEM 573Enzyme Kinetics N/A:...BIOCHEM 574Catalysis
N/A:...BIOCHEM 574Catalysis N/A:...Neuroscience and Cognitive Psychology [10]PSYC 2x1General Psychology
N/A:...Neuroscience and Cognitive Psychology [10]PSYC 2x1General Psychology N/A:...BIOPHYS 417Cellular Neurophysiology
N/A:...BIOPHYS 417Cellular Neurophysiology N/A:...PSYC 594Adaptive Systems
N/A:...PSYC 594Adaptive Systems N/A:...PSYC 600Psychology Graduate ProseminarNEUROSCI 615NeuroBiology of LearningNEUROSCI 616Cognition and Integration
N/A:...PSYC 600Psychology Graduate ProseminarNEUROSCI 615NeuroBiology of LearningNEUROSCI 616Cognition and Integration N/A:...PSYC 619Master's Thesis Project
N/A:...PSYC 619Master's Thesis Project N/A:...PSYC 640Neural Models
N/A:...PSYC 640Neural Models N/A:...PSYC 653Personality Psychology
N/A:...PSYC 653Personality Psychology N/A:...PSYC 808Cognitive Modeling
N/A:...PSYC 808Cognitive Modeling N/A:...Physics and Chaos Theory [10]PHYS 140General Physics I
N/A:...Physics and Chaos Theory [10]PHYS 140General Physics I N/A:...PHYS 141General Physics I Lab
N/A:...PHYS 141General Physics I Lab N/A:...PHYS 240General Physics II
N/A:...PHYS 240General Physics II N/A:...PHYS 241General Physics II Lab
N/A:...PHYS 241General Physics II Lab N/A:...PHYS 390Quantum Mechanics & Particle Physics
N/A:...PHYS 390Quantum Mechanics & Particle Physics N/A:...PHYS 413Physics of Nonlinear Dynamical Systems
N/A:...PHYS 413Physics of Nonlinear Dynamical Systems N/A:...APHYS 514Applied Physics Grad Seminar
N/A:...APHYS 514Applied Physics Grad Seminar N/A:...PSCS 520Empirical analysis of Non-Linear Systems
N/A:...PSCS 520Empirical analysis of Non-Linear Systems N/A:...PSCS 530Computer Modeling
N/A:...PSCS 530Computer Modeling N/A:The purpose of this course is to introduce students to the basic concepts, tools and issues which arise when using computers to model complex (adaptive) systems (CAS). The emphasis will be on agent-based, bottom-up computer models. (We will only briefly look at other approaches.) The bulk of the course will involve "learning by example", i.e., students will:PSCS 541Dynamical Complex Systems
N/A:The purpose of this course is to introduce students to the basic concepts, tools and issues which arise when using computers to model complex (adaptive) systems (CAS). The emphasis will be on agent-based, bottom-up computer models. (We will only briefly look at other approaches.) The bulk of the course will involve "learning by example", i.e., students will:PSCS 541Dynamical Complex Systems PSCS 541 - Introduction to Nonlinear Dynamics and the Physics of Complexity:An introduction to nonlinear science with an elementary treatment from the point of view of the physics of chaos and fractal growth
PSCS 541 - Introduction to Nonlinear Dynamics and the Physics of Complexity:An introduction to nonlinear science with an elementary treatment from the point of view of the physics of chaos and fractal growth
Timeline: Industry and Science Labs: My Employment History [13]RESEARCH
JOBSINDUSTRY
JOBSPublications541211111642TEACHINGEECS370EECS370EECS270,280ACADEMICSBSE in CSMS in CSPhD QualsPhD in Computer Engineering, Artificial IntelligenceMS in Aerospace Eng.MS in BioChemistry(95% MS Psychology)CGS in Complex SystemsMS in BioMed Eng.(85% MS Philosophy, MS EE)References [4] (available on reqest) Reference Name:...Titles...
Reference Name:...Titles...
...
...Additional Information...Address:...
...Phone:...E-mail:... Reference Name:...Titles...
Reference Name:...Titles...
...
...Additional Information...Address:...
...Phone:...E-mail:... Reference Name:...Titles...
Reference Name:...Titles...
...
...Additional Information...Address:...
...Phone:...E-mail:... Reference Name:...Titles...
Reference Name:...Titles...
...
...Additional Information...Address:...
...Phone:...E-mail:... - 2011 Lab.Chip








 I am currently a Software Engineer at Google and have 10+ years of Industry Experience as a Science Consultant,
I am currently a Software Engineer at Google and have 10+ years of Industry Experience as a Science Consultant,