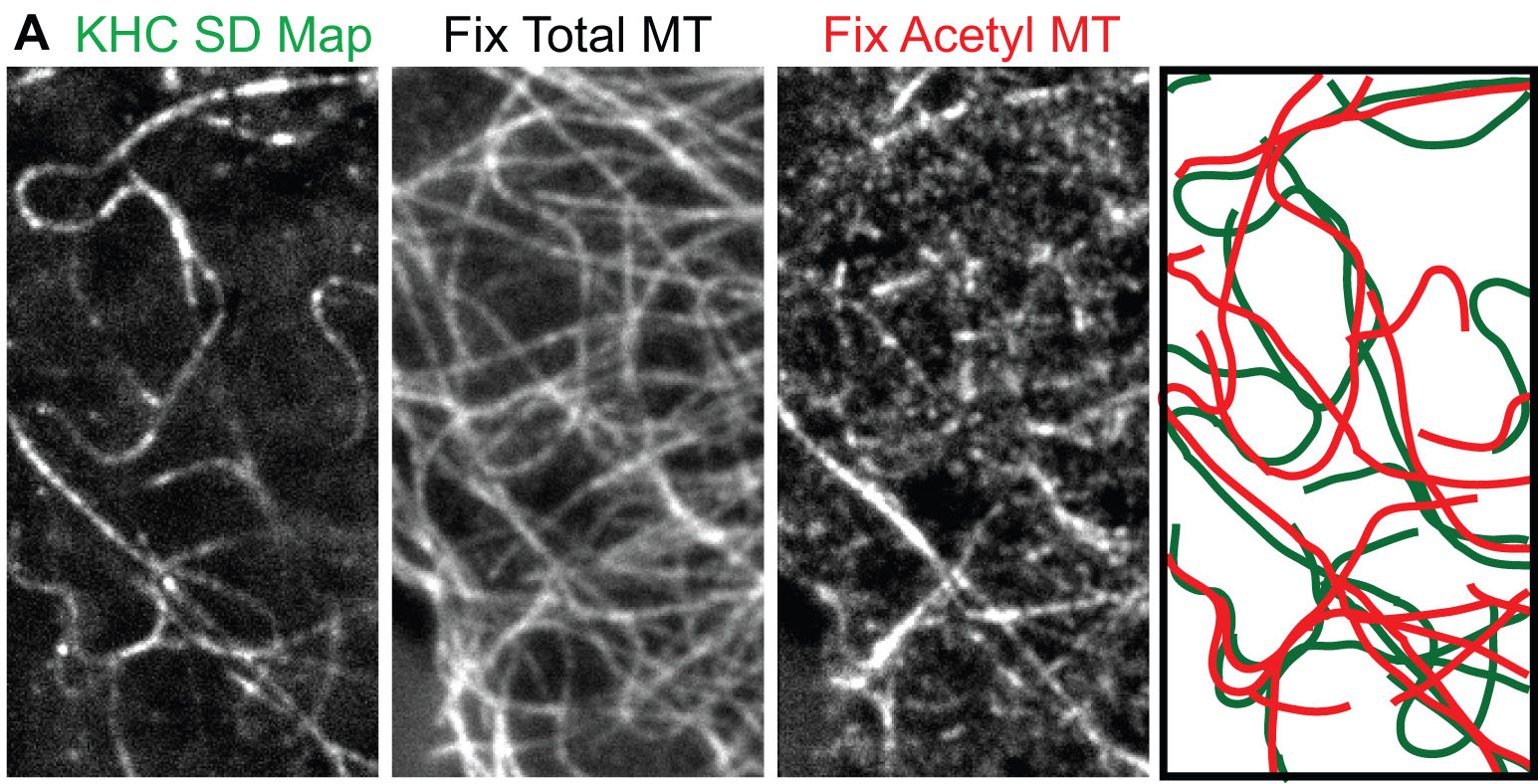
B. How kinesin motors respond to microtubule heterogeneity in cells.
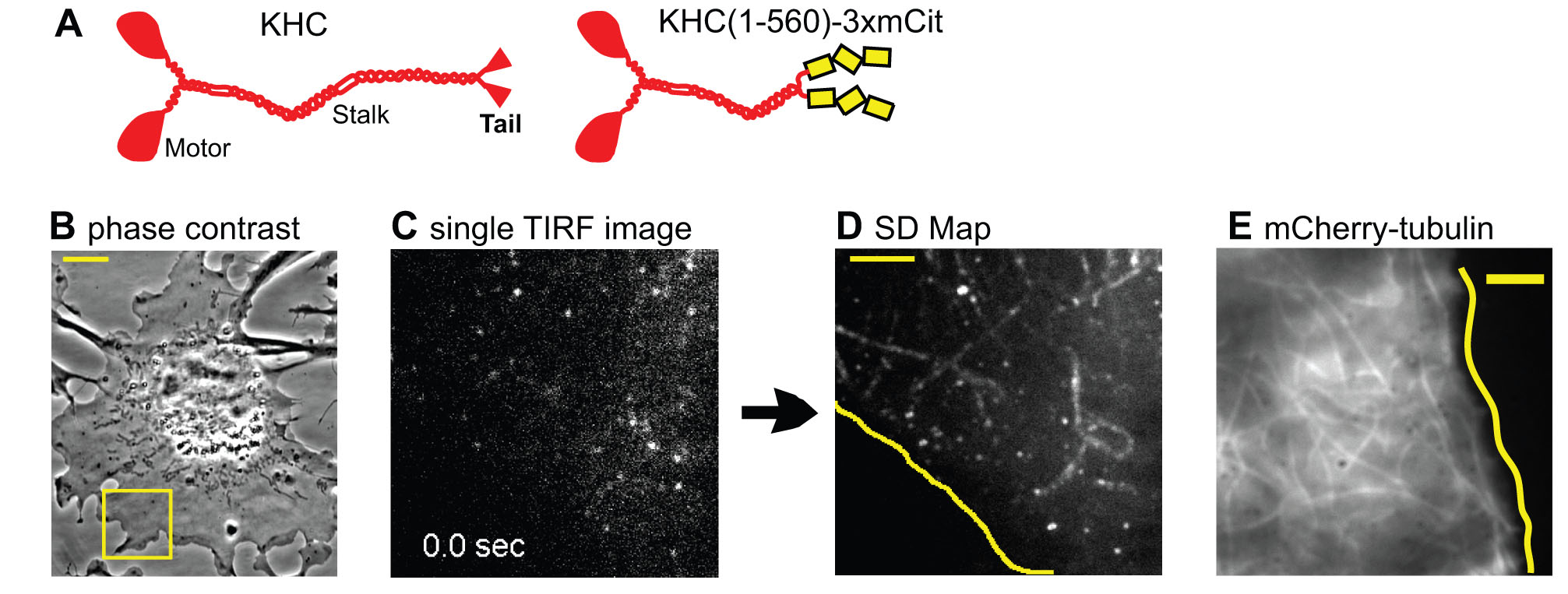
To investigate the behavior of kinesin motors in cells, we developed methods to track motors at the single molecule level in live cells. Truncated active motors are tagged with a three tandem FP tag (3xmCit), expressed in COS cells, and imaged by total internal reflection fluorescence (TIRF) microscopy (Movie 5). Post-image processing takes the ensemble of motility events that occurred during the image series and sums them into one image, the SD Map. Only a few microtubules are highlighted in the SD Map for Kinesin-1 (D) whereas imaging mCherry-tubulin by TIRF microscopy reveals many available microtubules (E):
Movie 5: Single molecule tracking by TIRF microscopy of KHC(1-891)-3xmCit motors expressed in a live COS cell. The image series was offline processed and color coded: green = single KHC(1-891)-3xmCit motors and red = microtubule tracks. The yellow line indicates the outline of the COS cell. The frame rate is 30 Hz. |
|||
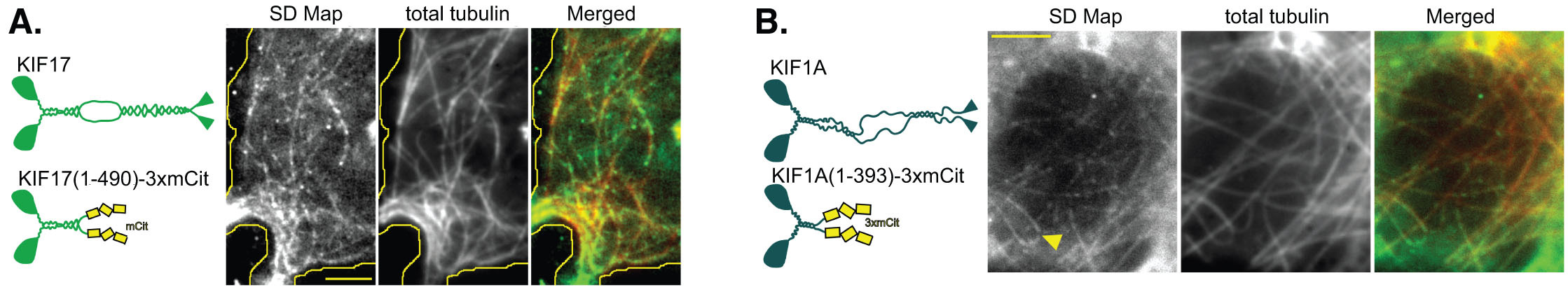
In contrast, single Kinesin-2 (KIF17) and Kinesin-3 (KIF1A) motors are not selective – their motility events occur on all available microtubules (below), including both stable and dynamic microtubules (not shown).
This work was the first to analyze the motility of kinesin motors in response to cellular microtubule heterogeneity. These studies reveal a new property by which kinesin families can be distinguished – the ability to select subpopulations of microtubules for preferred transport. In addition, our results have important implications for membrane trafficking in cells as the segregation of kinesin motors between subsets of microtubules enables the cell to target different trafficking events to specific cellular destinations.
This work suggests that the PTMs can provide a “tubulin code” for directing events along the microtubule polymer, analogous to the “histone code” that spatially regulates transcriptional events along chromatin. We are currently pursuing two questions about kinesin motors and microtubule PTMs:
1. What is the mechanistic basis by which Kinesin-1 distinguishes PTM-marked microtubules? To answer this question, we are using purified microtubules with defined PTMs and analyzing the effects on motor-microtubule binding, motor speed and motor processivity.
2. Do microtubule PTMs provide “road signs” to direct polarized kinesin transport in neuronal cells? To answer this question, we are using primary hippocampal neurons in which we alter PTMs and analyze the effects on motor and cargo trafficking events.
Relevant publications:
Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. 2006. Microtubule acetylation promotes Kinesin-1 binding and transport. Curr Biol. 16: 2166-2172. PMID: 17084703.
Cai D, Verhey KJ, Meyhofer E. 2007. Tracking Single Kinesin Molecules in the Cytoplasm of Mammalian Cells. Biophysical Journal 92: 4137-4144. PMID: 17400704.
Verhey KJ, Gaertig J. 2007. The tubulin code. Cell Cycle 6:2152-2160. PMID: 17786050. (review)
Hammond JW, Cai D, Verhey KJ. 2008. Tubulin modifications and their cellular functions. Curr Opin Cell Biol 20:71-76. PMID: 18226514. (review)
Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. 2009. Single Molecule Imaging Reveals Differences in Microtubule Track Selection Between Kinesin Motors. PLoS Biology 7: e1000216.