| Comparison: Ratio-based versus Significance-based Gene Selection Criteria | ||
| Back
to array experiments |
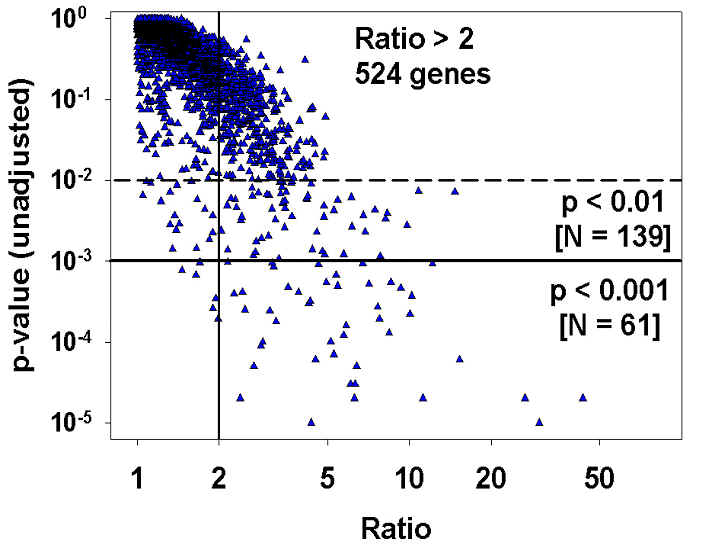
The experiment involved assessment of liver gene expression in eight young dwarf mice to eight controls. Each point shows a different gene; there are 2352 genes shown in all. There are 524 genes for which the ratio (dwarf/normal or normal/dwarf) exceeds 2-fold, but most of these are "false positives" in the sense that they do not even meet the convention p < 0.05 standard. Because a study looking at 2352 different genes would be expected to find 118 genes at p < 0.05 even if there were no real effects at all, a more cautious approach is to select p < 0.01 (23 false positives expected) or p < 0.001 (2 - 3 false positives). These two criteria would produce lists of 139 or 61 genes, respectively, including a number that would be missed by the ratio-based criterion. The figure is based upon data from Dozmorov, Galecki, Chang, Krzesicki, Vergara and Miller, "Gene expression profile of long-lived Snell dwarf mice," J. Gerontol. Biol. Sci., in press.
|