Ocean Viruses
Viral community omics, viruses and the biological pump, phage-host model systems, microscopy techniques
Viral community omics, viruses and the biological pump, phage-host model systems, microscopy techniques
Viral community omics in the Great Lakes; freshwater reservoirs of antibiotic resistance
Community structure and genomics of plastic biofilms in freshwater and oceans
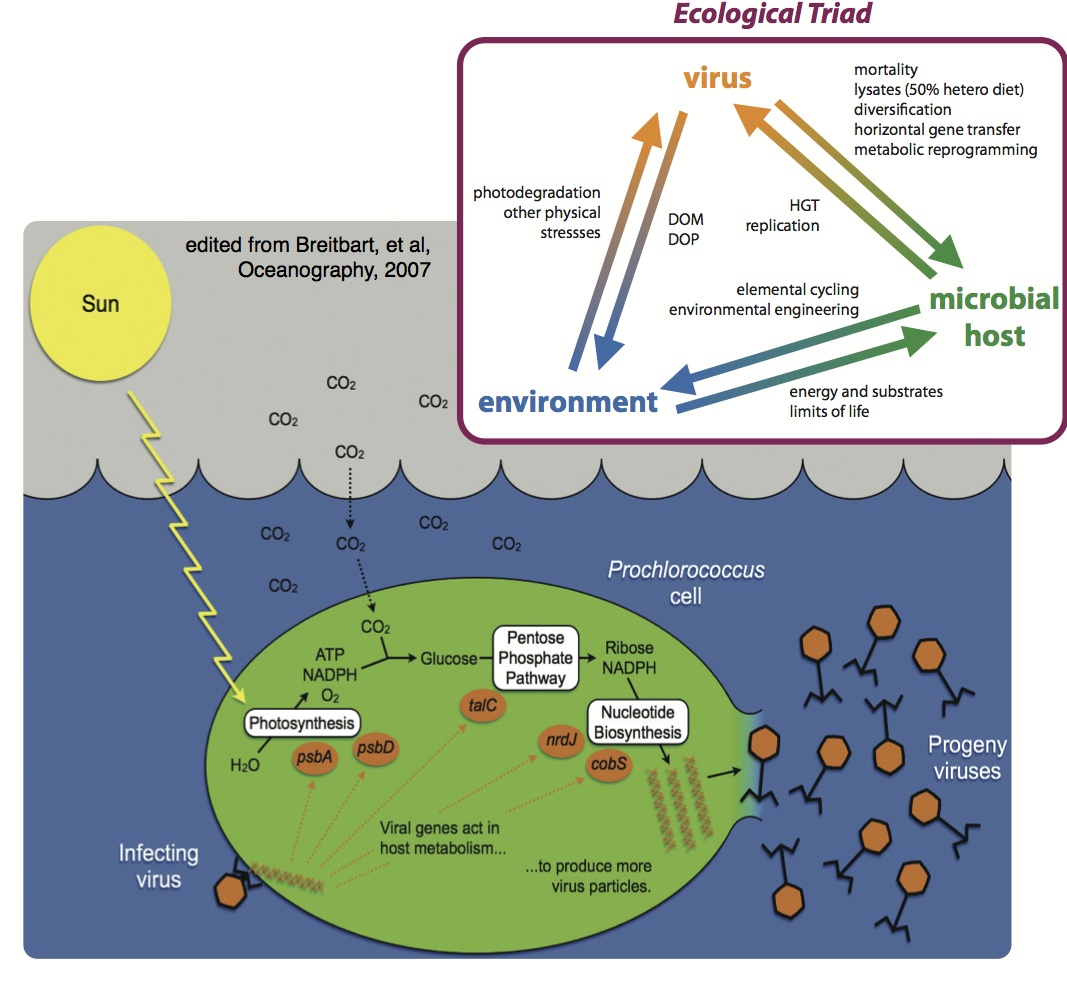 Ocean microbes produce half of the oxygen in the air we breathe and drive globally important biogeochemical cycles [1]. Their viruses modulate these cycles through mortality, horizontal gene transfer, and metabolic reprogramming [2]. About 1/3 of ocean microbes are infected by viruses per day [3], but we know little about these lysis products and their impacts on ecosystem function. This knowledge is critical for global carbon cycle modeling as viral lysis represents the greatest flux of carbon in the oceans (150 Gt/yr), dwarfing any other flux by >5-fold [3].
Ocean microbes produce half of the oxygen in the air we breathe and drive globally important biogeochemical cycles [1]. Their viruses modulate these cycles through mortality, horizontal gene transfer, and metabolic reprogramming [2]. About 1/3 of ocean microbes are infected by viruses per day [3], but we know little about these lysis products and their impacts on ecosystem function. This knowledge is critical for global carbon cycle modeling as viral lysis represents the greatest flux of carbon in the oceans (150 Gt/yr), dwarfing any other flux by >5-fold [3].
Infected microbes are metabolically reprogrammed towards states that enhance viral production, but likely alter microbial metabolic outputs. Virus are known to encode genes involved in photosynthesis [4, 5], nearly all of central carbon metabolism [6], nitrogen [7], and sulfur cycling [8]. However, data describing the the ramifications of these infections on ecosystem functions are lacking.
We seek to elucidate connections between biological oscillators (e.g., viruses and their hosts), metabolites, pathways, gene and protein regulation and global biogeochemical cycles, striving to predictively model the intracellular dynamics of phage-host-nutrient interactions and inform state-of-the-art multitrophic ecosystem models that now consider the role of viruses [9].
Genome-enabled investigations of (1) the influence of nutrient limitation during infection, (2) phage structural proteomics, (3) host range determinants, (4) experimental phage-host eco-evolutionary dynamics In collaboration with Christel Hassler (UNIGE), we study the viruses of the Southern Ocean using community-wide viral and microbial omics, culturing, flow cytometry, and physiochemical data from samples collected during the 2017 Antarctic Circumpolar Expedition (ACE). Samples span surface waters to 4000 m depth, continental margins to open water, and low to high productivity regimes.
In addition to novel viral and microbial biodiversity discovery in this near-unexplored ocean, the longterm goal is to develop an integrated virus-microbe interaction framework to better inform the drivers of microbial ecosystem functions in the Southern Ocean.
Development of high-resolution quantitative microscopy techniques to track viral infections. In collaboration with EMSL through the JGI-EMSL Collaborative Science Initiative (JECSI) and with Dr. Cristina Moraru (Uni Oldenburg).
In contrast with their ocean counterparts, the viral components of freshwater microbial communities barely have been explored. In addition to omics-driven novel biodiversity discovery, filling the knowledge gaps regarding how freshwater virus-host-environment interactions (above) differ from those in ocean systems will inform our perspective of carbon cycling in freshwater systems.
We study viruses infecting this harmful bloom forming algae through culturing, genomics, metagenomics, microscopy, and production assays. Our goal is (1) to better understand the impact of viral infection on the turnover of Microcystis populations through the bloom season and (2) to inform bloom ecology by modeling virus-host coevolution.
Collaborative project funded by the University of Michigan Water Center.
As the largest contiguous freshwater body on the planet, holding one fifth of the world's surface freshwater, the Great Lakes are one of the last unexplored frontiers in aquatic viral ecology. Using both viral and microbial omics datasets, we study the diversity and biogeography of Great Lakes virus populations, as well as the impact of virus-host interaction on freshwater ecosystem functions.
Collaborative project with the Denef Lab, with funding through the JGI CSP.
In collaboration with UM's Environmental Biotechnology Group (Wigginton, Raskin), we are developing methods for the high-throughput quantification and classification of antibiotic resistance genes in freshwater, farm, and other built environmental systems.
We study microscale interactions at the plastic interface (plastic-microbe-toxin) to better understand the fate of "one of the most ubiquitous and long-lasting recent changes to the surface of our planet" [1]: the aquatic plastic biome.
We are exploring the biogeography and functional potential of microbial communities living in biofilms on plastic debris in ocean and Great Lakes systems. We are largely motivated by questions concerning the environmental and public health impacts incurred from plastic-associated microbes, particularly concerning their propensity as an unexplored reservoir of pathogens and antibiotic resistance genes.
Oberbeckmann S, Osborn AM, Duhaime MB. 2016. Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris PLoS One.
We've taken a cross-disciplinary approach to define the ecological and environmental health risks of plastics in the Great Lakes. This includes field-based collections, developing methods for high-throughput, low-error quantification, improving circulation models to better predict transport, quantifying and qualifying the degree of plastic-bound organic pollutants, exploring plastic-dwelling microbial community dynamics, assessing the degree of plastic ingestion in key organisms, and contributing to national remediation action plans.
Cable RN, Beletsky D, Beletsky R, Wigginton K, Locke BW, Duhaime MB. 2017. Distribution and Modeled Transport of Plastic Pollution in the Great Lakes, the World's Largest Freshwater Resource. Frontiers in Environmental Science HTML
JM Hankett, WR Collin, P Yang, Z Chen, Duhaime MB. 2016. Low-volatility model demonstrates humidity affects environmental toxin deposition on plastics at a molecular level. Environmental Science & Technology 10.1021/acs.est.5b05598
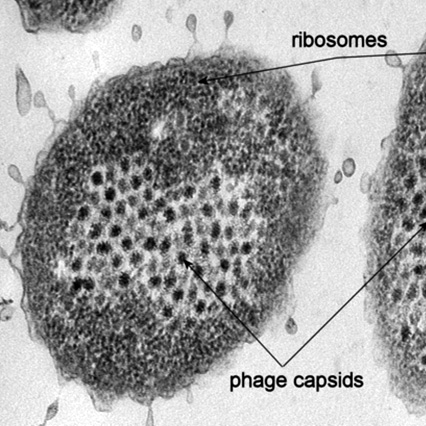
Ocean virus model system development: Heterotroph Pseudoalteromonas spp. and their phages
With funding from the JGI-EMSL Collaborative Science Initiative (JECSI) project "Building the phage-host-environment interaction data to scale from genes-to-ecosystems: Towards predictive modeling of wild microbial and viral community dynamics."
Duhaime MB, Wichels A, Waldmann J, Teeling H, Glöckner FO (2010). Ecogenomics and Genome Landscapes of Marine Pseudoalteromonas Phage H105/1. ISME Journal 5: 107-121.
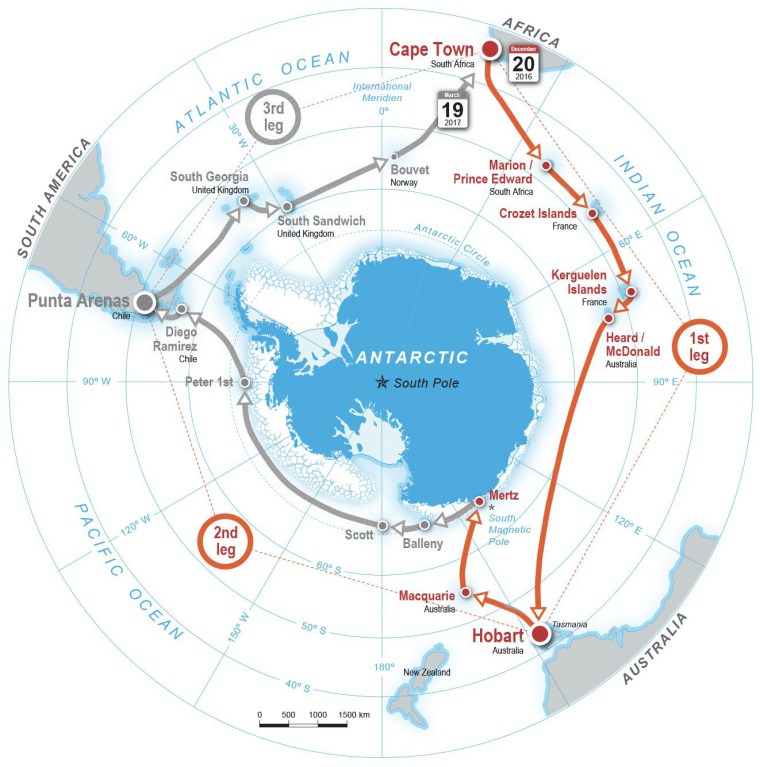
Southern Ocean Viruses
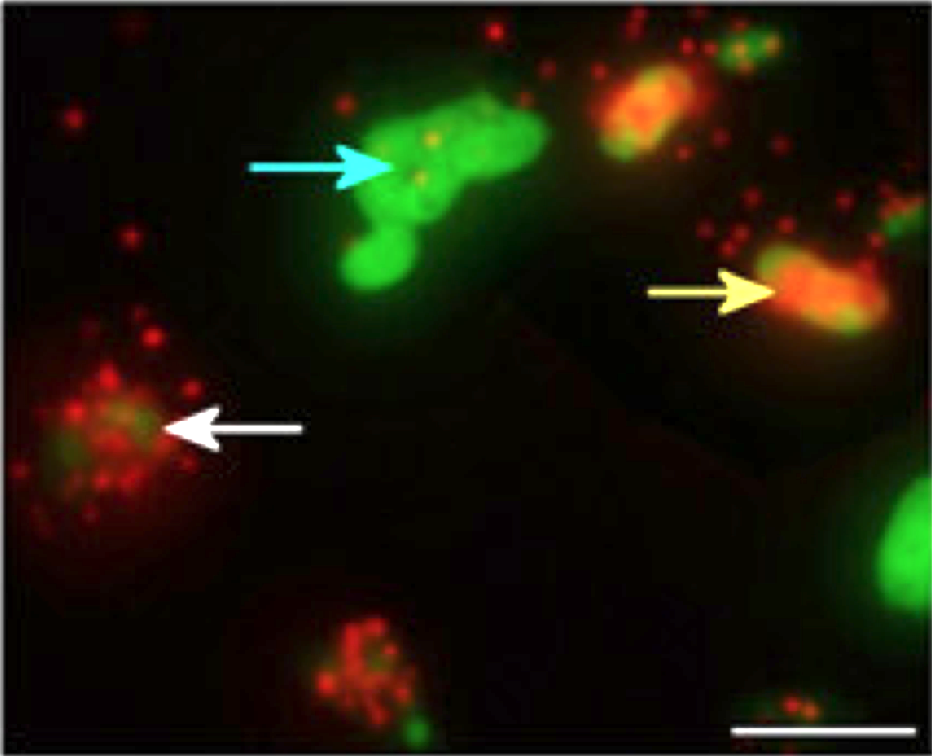
Tracking viral infections using phageFISH
Allers E, Moraru C, Duhaime MB, Beneze E, Solonenko N, Canosa JB, Amann R, MB Sullivan. (2013) Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environmental Microbiology
Freshwater Viruses
Current Lab Projects
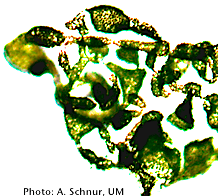
Viruses infecting Great Lakes Microcystis aeruginosa populations
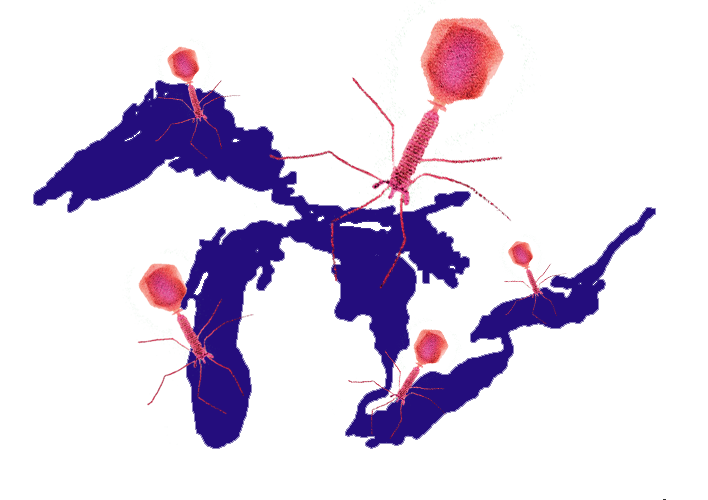
Great Lakes Viral Ecology
Quantitative metagenomics to evaluate reservoirs of antibiotic resistance
Plastic Microbiome
 Reports of the Great Pacific Garbage Patch, a higher-than-"normal" concentration soup of human-generated plastic debris in the North Pacific gyre, have brought fervent public awareness to the plastics in our oceans over the last decade. There is no corner left untouched by the presence of this foreign substrate in our oceans. New studies show the concentrations of plastic in our Great Lakes to be at times even higher than counts in the ocean.
Reports of the Great Pacific Garbage Patch, a higher-than-"normal" concentration soup of human-generated plastic debris in the North Pacific gyre, have brought fervent public awareness to the plastics in our oceans over the last decade. There is no corner left untouched by the presence of this foreign substrate in our oceans. New studies show the concentrations of plastic in our Great Lakes to be at times even higher than counts in the ocean.
Current Lab Projects

Plastic-associated microbial communities

Microplastics in the Great Lakes
This was a collaborative project funded by the University of Michigan Water Center.